3VAE
 
 | | Crystal Structure of M. tuberculosis LD-transpeptidase type 2 with Modified Catalytic Cysteine (C354) | | Descriptor: | DI(HYDROXYETHYL)ETHER, LD-transpeptidase type 2 | | Authors: | Erdemli, S, Bianchet, M.A, Gupta, R, Lamichhane, G, Amzel, L.M. | | Deposit date: | 2011-12-29 | | Release date: | 2012-12-12 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeting the Cell Wall of Mycobacterium tuberculosis: Structure and Mechanism of L,D-Transpeptidase 2.
Structure, 20, 2012
|
|
3TUR
 
 | | Crystal Structure of M. tuberculosis LD-transpeptidase type 2 complexed with a peptidoglycan fragment | | Descriptor: | 6-CARBOXYLYSINE, D-GLUTAMIC ACID, Di-mu-iodobis(ethylenediamine)diplatinum(II), ... | | Authors: | Bianchet, M.A, Erdemli, S.B, Gupta, R, Lamichhane, G, Amzel, L.M. | | Deposit date: | 2011-09-17 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Targeting the Cell Wall of Mycobacterium tuberculosis: Structure and Mechanism of L,D-Transpeptidase 2.
Structure, 20, 2012
|
|
3U1Q
 
 | | Crystal Structure of M. tuberculosis LD-transpeptidase type 2 with 2-Mercaptoethanol | | Descriptor: | BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, Mycobacteria Tuberculosis LD-transpeptidase type 2 | | Authors: | Erdemli, S, Bianchet, M.A, Gupta, R, Lamichhane, G, Amzel, L.M. | | Deposit date: | 2011-09-30 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of Mycobacterium tuberculosis L,D-transpeptidase 2 provides insights into targeting the cell wall of persisters
to be published
|
|
5D7H
 
 | | X-RAY CRYSTAL STRUCTURE OF L,D TRANSPEPTIDASE 2 FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | GLYCEROL, L,D-transpeptidase 2, SULFATE ION | | Authors: | Saavedra, H, Basta, L.A, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-08-13 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2 by biapenem and tebipenem.
BMC Biochem., 18, 2017
|
|
5DUJ
 
 | | Crystal structure of ldtMt2 in complex with Faropenem adduct | | Descriptor: | (2R,5R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydro-1,3-thiazole-4-carboxylic acid, BETA-MERCAPTOETHANOL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5DU7
 
 | |
5E1I
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T210 | | Descriptor: | (2S,3R,4R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-(methylsulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, GLYCEROL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2015-09-29 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5DZJ
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T206 in conformation A | | Descriptor: | (2~{R},3~{R},4~{R})-4-methyl-3-(2-oxidanylidene-2-propoxy-ethyl)sulfanyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, L,D-transpeptidase 2 | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5E51
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 1 with Faropenem adduct | | Descriptor: | (3R)-3-hydroxybutanal, L,D-transpeptidase 1 | | Authors: | Kumar, P, Lamichhane, G, Ginell, S.L. | | Deposit date: | 2015-10-07 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5DVP
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with Doripenem adduct | | Descriptor: | (2S,3R,4S)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, L,D-transpeptidase 2, PHOSPHONOACETALDEHYDE, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2015-09-21 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5E1G
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T208 | | Descriptor: | (2~{S},3~{R},4~{R})-4-(2-cyclohexyloxy-2-oxidanylidene-ethyl)sulfanyl-3-methyl-2-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, L,D-transpeptidase 2 | | Authors: | Kumar, P, Lamichhane, G, Ginell, S.L. | | Deposit date: | 2015-09-29 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5UWV
 
 | | Crystal structure of Mycobacterium abscessus L,D-transpeptidase 2 | | Descriptor: | L,D-TRANSPEPTIDASE 2 | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2017-02-21 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Mycobacterium abscessus l,d-Transpeptidases Are Susceptible to Inactivation by Carbapenems and Cephalosporins but Not Penicillins.
Antimicrob. Agents Chemother., 61, 2017
|
|
8B2Y
 
 | | Structure of the weakly red fluorescent protein csiFP4 from Clytia simplex | | Descriptor: | CHLORIDE ION, CsiFP4 chain A, SULFATE ION | | Authors: | Depernet, H, Engilberge, S, Lambert, G, Gotthard, G, Shaner, N, Royant, A. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure of the weakly red fluorescent protein csiFP4 from Clytia simplex
To Be Published
|
|
7F8P
 
 | | Crystal structure of the Mycobacterium tuberculosis L,D-transpeptidase-2 (LdtMt2) with new carbapenem drug T203 | | Descriptor: | (2R,3R)-3-methyl-4-(2-oxidanylidene-2-propan-2-yloxy-ethyl)sulfanyl-2-[(2R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]-2,3-dihydro-1H-pyrrole-5-carboxylic acid, (4R,5S,6S)-6-((R)-1-hydroxyethyl)-3-((2-isopropoxy-2-oxoethyl)thio)-4-methyl-7-oxo-1-azabicyclo[3.2.0]hept-2-ene-2-carboxylic acid, GLUTAMIC ACID, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2021-07-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Allosteric cooperation in beta-lactam binding to a non-classical transpeptidase.
Elife, 11, 2022
|
|
7F71
 
 | | Crystal structure of the Mycobacterium tuberculosis L,D-transpeptidase-2 (LdtMt2) with peptidoglycan sugar moiety and glutamate | | Descriptor: | GLUTAMIC ACID, GLYCEROL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2021-06-26 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Allosteric cooperation in beta-lactam binding to a non-classical transpeptidase.
Elife, 11, 2022
|
|
6DGM
 
 | | Streptococcus pyogenes phosphoglycerol transferase GacH in complex with sn-glycerol-1-phosphate | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Phosphoglycerol transferase GacH, ... | | Authors: | Edgar, R.J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2018-05-17 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of glycerol phosphate modification on streptococcal rhamnose polysaccharides.
Nat.Chem.Biol., 15, 2019
|
|
6DQ3
 
 | | Streptococcus pyogenes deacetylase PplD in complex with acetate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IMIDAZOLE, ... | | Authors: | Li, J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2018-06-10 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | PplD is a de-N-acetylase of the cell wall linkage unit of streptococcal rhamnopolysaccharides
Nat Commun, 13, 2022
|
|
6OUG
 
 | | Structure of drug-resistant V27A mutant of the influenza M2 proton channel bound to spiroadamantyl amine inhibitor, TM + cytosolic helix construct | | Descriptor: | (1r,1'S,3'S,5'S,7'S)-spiro[cyclohexane-1,2'-tricyclo[3.3.1.1~3,7~]decan]-4-amine, Matrix protein 2 | | Authors: | Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2019-05-04 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | X-ray Crystal Structures of the Influenza M2 Proton Channel Drug-Resistant V27A Mutant Bound to a Spiro-Adamantyl Amine Inhibitor Reveal the Mechanism of Adamantane Resistance.
Biochemistry, 59, 2020
|
|
7ZH2
 
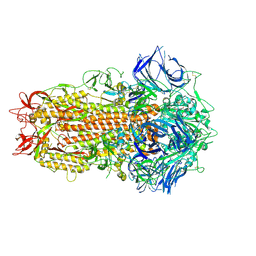 | | SARS CoV Spike protein, Closed C1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
7ZH5
 
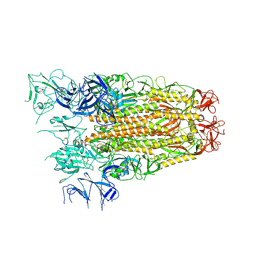 | | SARS CoV Spike protein, Open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
7ZH1
 
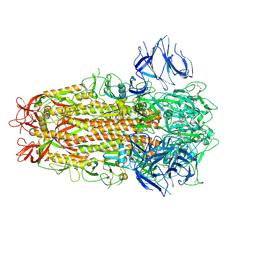 | | SARS CoV Spike protein, Closed C3 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
5AK2
 
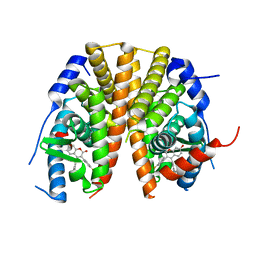 | | Oxyphenylpropenoic acids as Oral Selective Estrogen Receptor Down- Regulators. | | Descriptor: | (E)-3-[4-[[3-(4-fluoranyl-2-methyl-phenyl)-7-oxidanyl-2-oxidanylidene-chromen-4-yl]methyl]phenyl]prop-2-enoic acid, ESTROGEN RECEPTOR | | Authors: | Degorce, S, Bailey, A, Callis, R, De Savi, C, Ducray, R, Lamot, P, MacFaul, P, Maudet, M, Norman, R.A, Scott, J.S, Phillips, C. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Investigation of (E)-3-[4-(2-Oxo-3-Aryl-Chromen-4-Yl)Oxyphenyl]Acrylic Acids as Oral Selective Estrogen Receptor Down-Regulators.
J.Med.Chem., 58, 2015
|
|
7O70
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(4R,7S)-12-chloro-14-fluoro-13-(2-fluoro-6-hydroxyphenyl)-4-methyl-10-oxa-2,5,16,18-tetrazatetracyclo[9.7.1.0^(2,7).0^(15,19)]nonadeca-1(18),11,13,15(19),16-pentaen-5-en-1-one-yl]prop-2, CALCIUM ION, GTPase KRas, ... | | Authors: | Phillips, C. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Discovery of AZD4625, a Covalent Allosteric Inhibitor of the Mutant GTPase KRAS G12C .
J.Med.Chem., 65, 2022
|
|
7O83
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(7S)-11-chloro-12-(5-methyl-1H-indazol-4-yl)-9-oxa-2,5,15,17-tetrazatetracyclo[8.7.1.02,7.014,18]octadeca-1(17),10,12,14(18),15-pentaen-5-yl]prop-2-en-1-one, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C, Breed, J. | | Deposit date: | 2021-04-14 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery of AZD4625, a Covalent Allosteric Inhibitor of the Mutant GTPase KRAS G12C .
J.Med.Chem., 65, 2022
|
|
6BOI
 
 | | Crystal Structure of LdtMt2 (56-408) with a panipenem adduct at the active site cysteine-354 | | Descriptor: | (3S,5R)-5-[(2R,3R)-1,3-dihydroxybutan-2-yl]-3-({(3R)-1-[(1E)-ethanimidoyl]pyrrolidin-3-yl}sulfanyl)-L-proline, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Saavedra, H, Bianchet, M.A. | | Deposit date: | 2017-11-20 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structures and Mechanism of Inhibition of Mycobacterium tuberculosis L,D-transpeptidase 2 by Panipenem
To Be Published
|
|
