3KJO
 
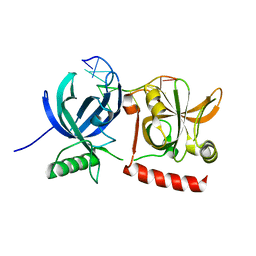 | | Crystal Structure of hPOT1V2-dTrUd(AGGGTTAG) | | Descriptor: | DNA/RNA (5'-D(*T)-R(P*U)-D(P*AP*GP*GP*GP*TP*TP*AP*G)-3'), Protection of telomeres protein 1 | | Authors: | Nandakumar, J, Cech, T.R, Podell, E.R. | | Deposit date: | 2009-11-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How telomeric protein POT1 avoids RNA to achieve specificity for single-stranded DNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2HVQ
 
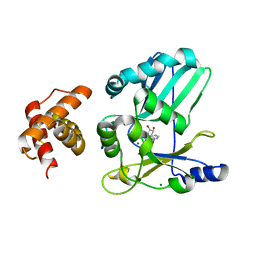 | | Structure of Adenylated full-length T4 RNA Ligase 2 | | Descriptor: | Hypothetical 37.6 kDa protein in Gp24-hoc intergenic region, MAGNESIUM ION | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
2HVS
 
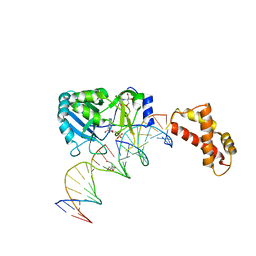 | | Structure of T4 RNA Ligase 2 with Nicked 5'-Adenylated nucleic acid duplex containing a 2'-deoxyribonucleotide at the nick | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*TP*TP*G)-3', 5'-D(*CP*AP*AP*TP*TP*GP*CP*GP*AP*C)-R(P*(OMC)P*C)-3', ... | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
2HVR
 
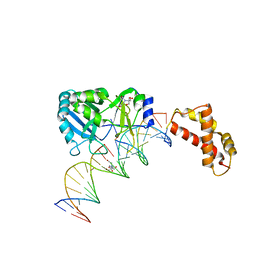 | | Structure of T4 RNA Ligase 2 with Nicked 5'-Adenylated nucleic acid duplex containing a 3'-deoxyribonucleotide at the nick | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*TP*TP*G)-3', 5'-D(*CP*AP*AP*TP*TP*GP*CP*GP*AP*C)-R(P*(OMC)P*C)-3', ... | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
6LU9
 
 | |
3U93
 
 | |
3U92
 
 | |
3U94
 
 | |
2DT2
 
 | | Crystal structure of the complex formed between goat signalling protein with pentasaccharide at 2.9A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1 | | Authors: | Ethayathulla, A.S, Kumar, J, Srivastava, D.B, Singh, N, Sharma, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Carbohydrate-binding properties of goat secretory glycoprotein (SPG-40) and its functional implications: structures of the native glycoprotein and its four complexes with chitin-like oligosaccharides
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
5WIR
 
 | | Structure of the TRF1-TERB1 interface | | Descriptor: | TERB1-TBM, Telomeric repeat-binding factor 1 | | Authors: | Nandakumar, J, Pendlebury, D.F, Smith, E.M, Tesmer, V.M. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dissecting the telomere-inner nuclear membrane interface formed in meiosis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8U8L
 
 | | X-ray crystal structure of TEBP-2 MCD3 with ds DNA | | Descriptor: | CESIUM ION, DNA (5'-D(*CP*TP*GP*TP*TP*AP*GP*GP*CP*TP*TP*AP*GP*GP*CP*TP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*GP*CP*CP*TP*AP*AP*GP*CP*CP*TP*AP*AP*CP*A)-3'), ... | | Authors: | Nandakumar, J, Padmanaban, S. | | Deposit date: | 2023-09-18 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caenorhabditis elegans telomere-binding proteins TEBP-1 and TEBP-2 adapt the Myb module to dimerize and bind telomeric DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U8M
 
 | | X-ray crystal structure of TEBP-1 MCD2 homodimer | | Descriptor: | COBALT (II) ION, Double-strand telomeric DNA-binding proteins 1 | | Authors: | Nandakumar, J, Padmanaban, S. | | Deposit date: | 2023-09-18 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Caenorhabditis elegans telomere-binding proteins TEBP-1 and TEBP-2 adapt the Myb module to dimerize and bind telomeric DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7S3L
 
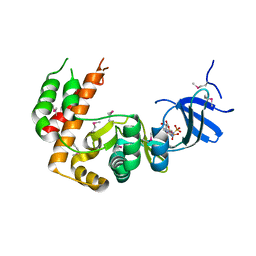 | |
5I2Y
 
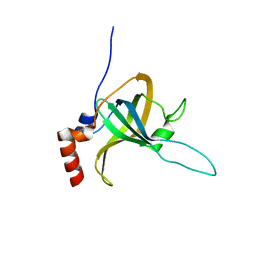 | | Crystal Structure of TPP1 K170A | | Descriptor: | Adrenocortical dysplasia protein homolog | | Authors: | Nandakumar, J, Smith, E.M. | | Deposit date: | 2016-02-09 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional consequences of a disease mutation in the telomere protein TPP1.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5I2X
 
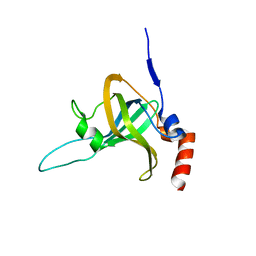 | | Crystal Structure of TPP1 K170del | | Descriptor: | Adrenocortical dysplasia protein homolog | | Authors: | Nandakumar, J, Smith, E. | | Deposit date: | 2016-02-09 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional consequences of a disease mutation in the telomere protein TPP1.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8SH0
 
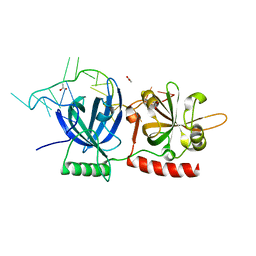 | |
8SH1
 
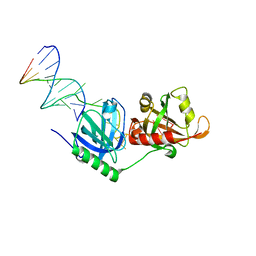 | |
7LDG
 
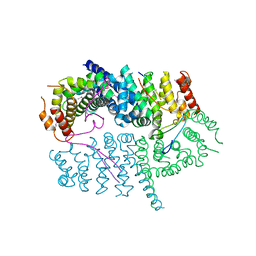 | | Crystal structure of the MEILB2-BRCA2 complex | | Descriptor: | Breast cancer type 2 susceptibility protein, Heat shock factor 2-binding protein | | Authors: | Nandakumar, J, Pendlebury, D.F. | | Deposit date: | 2021-01-13 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of a meiosis-specific complex central to BRCA2 localization at recombination sites.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4UQQ
 
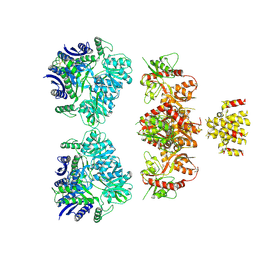 | | Electron density map of GluK2 desensitized state in complex with 2S,4R-4-methylglutamate | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, KAINATE 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQJ
 
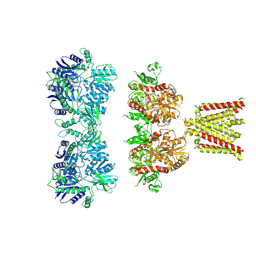 | | Cryo-EM density map of GluA2em in complex with ZK200775 | | Descriptor: | GLUTAMATE RECEPTOR 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQK
 
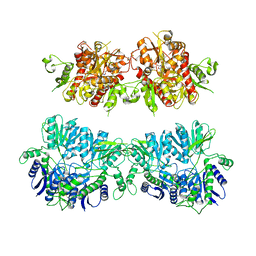 | | Electron density map of GluA2em in complex with quisqualate and LY451646 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2 | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
2B31
 
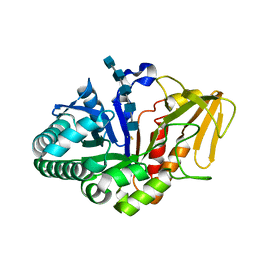 | | Crystal structure of the complex formed between goat signalling protein with pentasaccharide at 3.1 A resolution reveals large scale conformational changes in the residues of TIM barrel | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, ... | | Authors: | Ethayathulla, A.S, Kumar, J, Srivastava, D.B, Singh, N, Sharma, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2005-09-19 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the complex formed between goat signalling protein with pentasaccharide at 3.1 A resolution reveals large scale conformational changes in the residues of TIM barrel
TO BE PUBLISHED
|
|
1SQY
 
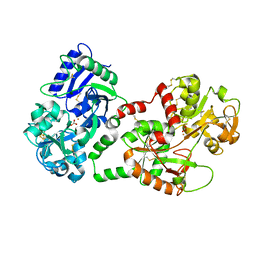 | | Structure of human diferric lactoferrin at 2.5A resolution using crystals grown at pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Vikram, P, Prem Kumar, R, Singh, N, Kumar, J, Ethayathulla, A.S, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2004-03-22 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of human diferric lactoferrin at 2.5A resolution using crystals grown at pH 6.5.
To be Published
|
|
4H3O
 
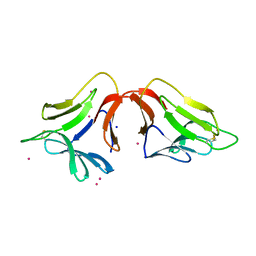 | | Crystal structure of a new form of lectin from Allium sativum at 2.17 A resolution | | Descriptor: | CADMIUM ION, Lectin, SODIUM ION | | Authors: | Kumar, S, Yamini, S, Kumar, J, Kaur, P, Singh, T.P, Dey, S. | | Deposit date: | 2012-09-14 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of a new form of lectin from Allium sativum at 2.17 A resolution
To be Published
|
|
7YSV
 
 | |
