4GOB
 
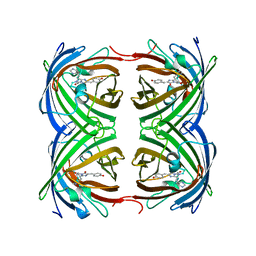 | | Low pH Crystal Structure of a reconstructed Kaede-type Red Fluorescent Protein, Least Evolved Ancestor (LEA) | | Descriptor: | Kaede-type Fluorescent Protein | | Authors: | Kim, H, Grunkemeyer, T.J, Chen, L, Fromme, R, Wachter, R.M. | | Deposit date: | 2012-08-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Acid-base catalysis and crystal structures of a least evolved ancestral GFP-like protein undergoing green-to-red photoconversion.
Biochemistry, 52, 2013
|
|
1BLL
 
 | |
1BPM
 
 | |
1GYP
 
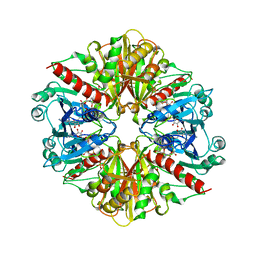 | | CRYSTAL STRUCTURE OF GLYCOSOMAL GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM LEISHMANIA MEXICANA: IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN AND A NEW POSITION FOR THE INORGANIC PHOSPHATE BINDING SITE | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Kim, H, Feil, I.K, Verlinde, C.L.M.J, Petra, P.H, Hol, W.G.J. | | Deposit date: | 1995-08-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Leishmania mexicana: implications for structure-based drug design and a new position for the inorganic phosphate binding site.
Biochemistry, 34, 1995
|
|
1J32
 
 | |
7BWK
 
 | | Structure of DotL(656-783)-IcmS-IcmW-LvgA-VpdB(461-590) derived from Legionella pneumophila | | Descriptor: | Hypothetical virulence protein, IcmO (DotL), IcmS, ... | | Authors: | Kim, H, Kwak, M.J, Oh, B.H. | | Deposit date: | 2020-04-14 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for effector protein recognition by the Dot/Icm Type IVB coupling protein complex.
Nat Commun, 11, 2020
|
|
7X15
 
 | | Crystal structure of MIGA2 LD targeting domain | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, FORMIC ACID, Mitoguardin 2 | | Authors: | Kim, H, Lee, C. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Structural basis for mitoguardin-2 mediated lipid transport at ER-mitochondrial membrane contact sites.
Nat Commun, 13, 2022
|
|
7X14
 
 | | Crystal structure of phospho-FFAT motif of MIGA2 bound to VAPB | | Descriptor: | MIGA2 phospho FFAT motif, SULFATE ION, Vesicle-associated membrane protein-associated protein B | | Authors: | Kim, H, Lee, C. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.675 Å) | | Cite: | Structural basis for mitoguardin-2 mediated lipid transport at ER-mitochondrial membrane contact sites.
Nat Commun, 13, 2022
|
|
7VWX
 
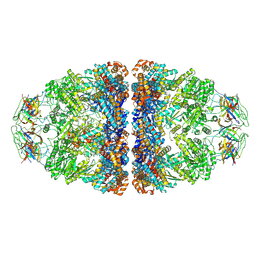 | | CryoEM structure of football-shaped GroEL:ES2 with RuBisCO | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, Ribulose bisphosphate carboxylase | | Authors: | Kim, H, Roh, S.H. | | Deposit date: | 2021-11-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Cryo-EM structures of GroEL:ES 2 with RuBisCO visualize molecular contacts of encapsulated substrates in a double-cage chaperonin.
Iscience, 25, 2022
|
|
5X90
 
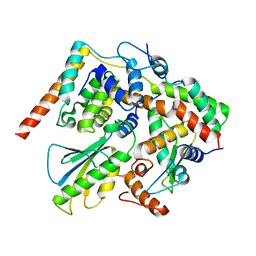 | | Structure of DotL(656-783)-IcmS-IcmW-LvgA derived from Legionella pneumophila | | Descriptor: | Hypothetical virulence protein, IcmO (DotL), IcmS, ... | | Authors: | Kim, H, Kwak, M.J, Kim, J.D, Kim, Y.G, Oh, B.H. | | Deposit date: | 2017-03-04 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Architecture of the type IV coupling protein complex of Legionella pneumophila
Nat Microbiol, 2, 2017
|
|
8GTG
 
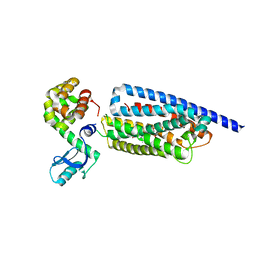 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-I-152 by XFEL | | Descriptor: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N},~{N}-bis(2-methoxyethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8GTI
 
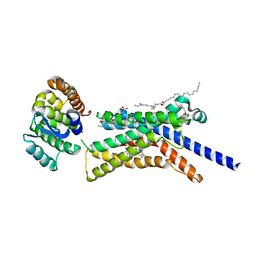 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C205 by XFEL | | Descriptor: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N}-butyl-~{N}-(cyclopropylmethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1, ... | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8GTM
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C203 by XFEL | | Descriptor: | 7-(4-bromanyl-2,6-dimethoxy-phenyl)-4,8-dimethyl-~{N},~{N}-bis[4,4,4-tris(fluoranyl)butyl]-1$l^{4},3,5,9-tetrazabicyclo[4.3.0]nona-1(6),2,4,8-tetraen-2-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8YHU
 
 | | hTLR3/minibinder 8.6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 3, ... | | Authors: | Kim, H, Kim, H. | | Deposit date: | 2024-02-28 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | De novo design of protein minibinder agonists of TLR3.
Nat Commun, 16, 2025
|
|
8YHT
 
 | | hTLR3/minibinder 7.7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 3, ... | | Authors: | Kim, H, Kim, H. | | Deposit date: | 2024-02-28 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | De novo design of protein minibinder agonists of TLR3.
Nat Commun, 16, 2025
|
|
5XHW
 
 | | Crystal structure of HddC from Yersinia pseudotuberculosis | | Descriptor: | Putative 6-deoxy-D-mannoheptose pathway protein, SULFATE ION | | Authors: | Park, J, Kim, H, Kim, S, Shin, D.H. | | Deposit date: | 2017-04-24 | | Release date: | 2018-04-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of d-glycero-alpha-d-manno-heptose-1-phosphate guanylyltransferase from Yersinia pseudotuberculosis.
Biochim. Biophys. Acta, 1866, 2018
|
|
4ZG0
 
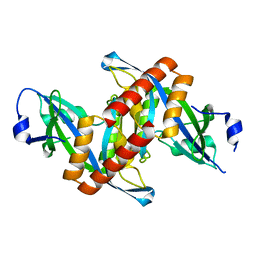 | | Crystal structure of Mouse Syndesmos protein | | Descriptor: | Protein syndesmos | | Authors: | Lee, I, Kim, H, Yoo, J, Cho, H, Lee, W. | | Deposit date: | 2015-04-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of syndesmos and its interaction with Syndecan-4 proteoglycan
Biochem.Biophys.Res.Commun., 463, 2015
|
|
4GCV
 
 | |
6NBA
 
 | | Crystal structure of Human Cystathionine gamma lyase with S-3-Carboxpropyl-L-Cysteine | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, Cystathionine gamma-lyase | | Authors: | Kim, H, Yadav, P.K, Banerjee, R, Cho, U.-S. | | Deposit date: | 2018-12-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | S-3-Carboxypropyl-l-cysteine specifically inhibits cystathionine gamma-lyase-dependent hydrogen sulfide synthesis.
J.Biol.Chem., 294, 2019
|
|
8ZKE
 
 | | Cryo-EM structure of inward-facing Anhydromuropeptide permease (AmpG) in complex with GlcNAc-1,6-anhMurNAc | | Descriptor: | (2R)-2-[[(1R,2S,3R,4R,5R)-4-acetamido-2-[(2S,3R,4R,5S,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy]propanoic acid, Muropeptide transporter | | Authors: | Chang, N, Kim, U, Yoo, Y, Kim, H, Cho, H. | | Deposit date: | 2024-05-16 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM structure of inward-facing Anhydromuropeptide permease (AmpG) in complex with GlcNAc-1,6-anhMurNAc
To Be Published
|
|
8ZBB
 
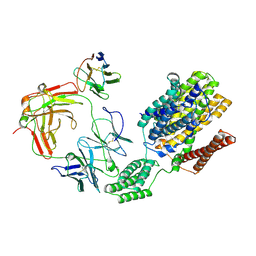 | | Cryo-EM structure of outward state Anhydromuropeptide permease (AmpG) G50W/L269W | | Descriptor: | Muropeptide transporter,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, anti-BRIL Fab Light chain, ... | | Authors: | Yoo, Y, Chang, N, Kim, U, Kim, H, Cho, H. | | Deposit date: | 2024-04-26 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Cryo-EM structure of outward state Anhydromuropeptide permease (AmpG) G50W/L269W
To Be Published
|
|
7N99
 
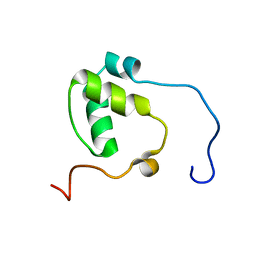 | | SDE2 SAP domain apo structure | | Descriptor: | Isoform 2 of Replication stress response regulator SDE2 | | Authors: | Paung, Y, Weinheimer, A.S, Rageul, J, Khan, A, Ho, B, Tong, M, Alphonse, S, Seeliger, M.A, Kim, H. | | Deposit date: | 2021-06-17 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Extended DNA-binding interfaces beyond the canonical SAP domain contribute to the function of replication stress regulator SDE2 at DNA replication forks.
J.Biol.Chem., 298, 2022
|
|
7FEQ
 
 | | Cryo-EM structure of apo BsClpP at pH 6.5 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7FES
 
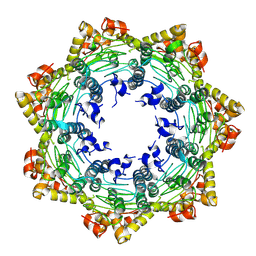 | | Cryo-EM structure of apo BsClpP at pH 4.2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7FEP
 
 | | Cryo-EM structure of BsClpP-ADEP1 complex at pH 6.5 | | Descriptor: | ADEP1, ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
