5JBI
 
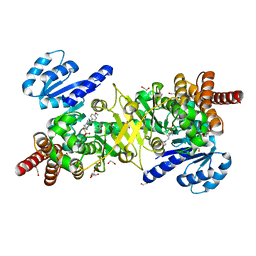 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC52 and manganese | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-04-13 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
5JO0
 
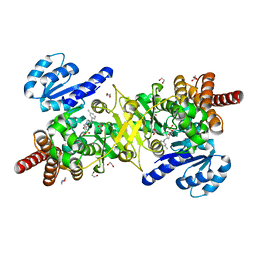 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC56 and manganese | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-05-01 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
5JNL
 
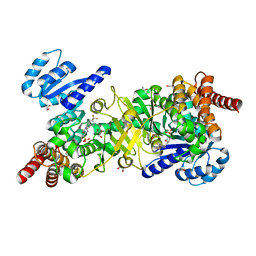 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC54 and manganese | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-04-30 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
5JC1
 
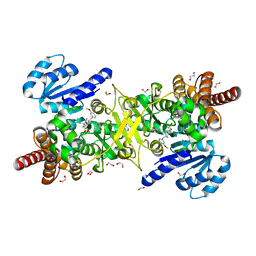 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC55 and manganese | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-04-14 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
8D36
 
 | |
8DAO
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV44-79 | | Descriptor: | COV44-79 heavy chain constant domain, COV44-79 heavy chain variable domain, COV44-79 light chain constant domain, ... | | Authors: | Lin, T.H, Lee, C.C.D, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-13 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8D6Z
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV91-27 | | Descriptor: | Neutralizing antibody COV91-27 heavy chain, Neutralizing antibody COV91-27 light chain, Spike protein S2 fusion peptide | | Authors: | Lee, C.C.D, Lin, T.H, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8DTX
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV89-22 | | Descriptor: | COV89-22 heavy chain, COV89-22 light chain, Spike protein S2' stem helix peptide | | Authors: | Lin, T.H, Lee, C.C.D, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTR
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV30-14 | | Descriptor: | COV30-14 heavy chain, COV30-14 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTT
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV93-03 | | Descriptor: | COV93-03 heavy chain, COV93-03 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
5U3I
 
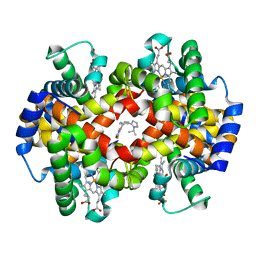 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN S (LIGANDED SICKLE CELL HEMOGLOBIN) COMPLEXED WITH GBT compound 31 | | Descriptor: | 2-methoxy-5-({2-[1-(propan-2-yl)-1H-pyrazol-5-yl]pyridin-3-yl}methoxy)pyridine-4-carbaldehyde, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Partridge, J.R, Choy, R.M, Li, Z, Metcalf, B. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of GBT440, an Orally Bioavailable R-State Stabilizer of Sickle Cell Hemoglobin.
ACS Med Chem Lett, 8, 2017
|
|
5UFJ
 
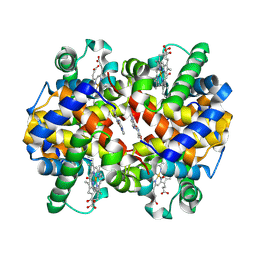 | | Crystal Structure of Carbonmonoxy Hemoglobin S (Liganded Sickle Cell Hemoglobin) Complexed with GBT Compound 6 | | Descriptor: | 5-[(imidazo[1,2-a]pyridin-8-yl)methoxy]-2-methoxypyridine-4-carbaldehyde, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Partridge, J.R, Choy, R.M, Li, Z, Metcalf, B. | | Deposit date: | 2017-01-04 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of GBT440, an Orally Bioavailable R-State Stabilizer of Sickle Cell Hemoglobin.
ACS Med Chem Lett, 8, 2017
|
|
5JSJ
 
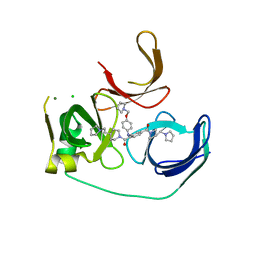 | | Crystal structure of Spindlin1 bound to compound EML631 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Spindlin-1, ... | | Authors: | Su, X, Li, H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Developing Spindlin1 small-molecule inhibitors by using protein microarrays
Nat. Chem. Biol., 13, 2017
|
|
5JSG
 
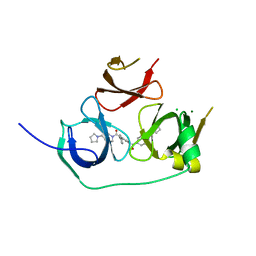 | | Crystal structure of Spindlin1 bound to compound EML405 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Spindlin-1, ... | | Authors: | Su, X, Li, H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Developing Spindlin1 small-molecule inhibitors by using protein microarrays
Nat. Chem. Biol., 13, 2017
|
|
6QDJ
 
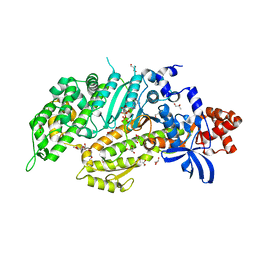 | | Molecular features of the UNC-45 chaperone critical for binding and folding muscle myosin | | Descriptor: | 1,4-BUTANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Meinhart, A, Clausen, T, Arnese, R. | | Deposit date: | 2019-01-02 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Molecular features of the UNC-45 chaperone critical for binding and folding muscle myosin.
Nat Commun, 10, 2019
|
|
6QDL
 
 | |
6QDM
 
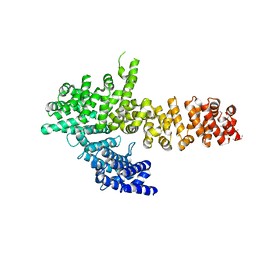 | |
6QDK
 
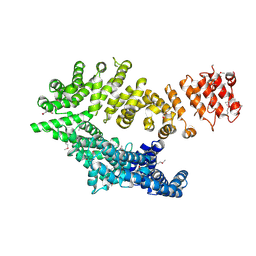 | |
7NPO
 
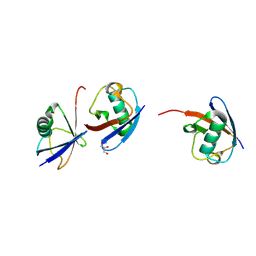 | | Branched K48-K63-Ub3 | | Descriptor: | GLYCEROL, Polyubiquitin-B | | Authors: | Lange, S.M, Kwasna, D, Kulathu, Y. | | Deposit date: | 2021-02-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | VCP/p97-associated proteins are binders and debranching enzymes of K48-K63-branched ubiquitin chains.
Nat.Struct.Mol.Biol., 2024
|
|
7NBB
 
 | |
6MDU
 
 | | Crystal structure of NDM-1 with compound 7 | | Descriptor: | 1,5-diphenyl-N-(1H-tetrazol-5-yl)-1H-pyrazole-3-carboxamide, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
6MD8
 
 | | Crystal structure of CTX-M-14 with compound 3 | | Descriptor: | 1-(2,4-dichlorophenyl)-4-(1H-tetrazol-5-yl)-1H-pyrazol-5-amine, Beta-lactamase CTX-M-14, DIMETHYL SULFOXIDE | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-04 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
6MEY
 
 | | Crystal structure of KPC-2 with compound 9 | | Descriptor: | (2R)-2-phenyl-2-(phenylamino)-N-(1H-tetrazol-5-yl)acetamide, ACETATE ION, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
6M7I
 
 | | Crystal structure of KPC-2 with compound 3 | | Descriptor: | 1-(2,4-dichlorophenyl)-4-(1H-tetrazol-5-yl)-1H-pyrazol-5-amine, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
6MIA
 
 | | Crystal structure of CTX-M-14 with compound 6 | | Descriptor: | 3-(1H-tetrazol-5-ylmethyl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one, Beta-lactamase | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
