4P8O
 
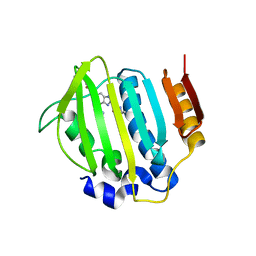 | | S. aureus gyrase bound to an aminobenzimidazole urea inhibitor | | Descriptor: | 1-ethyl-3-[5-(5-fluoropyridin-3-yl)-7-(pyrimidin-2-yl)-1H-benzimidazol-2-yl]urea, DNA gyrase subunit B | | Authors: | Jacobs, M.D. | | Deposit date: | 2014-03-31 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Second-generation antibacterial benzimidazole ureas: discovery of a preclinical candidate with reduced metabolic liability.
J.Med.Chem., 57, 2014
|
|
3BGZ
 
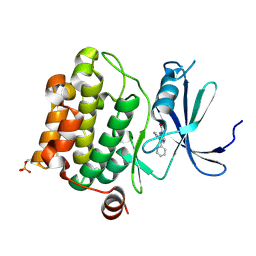 | | Human Pim-1 kinase in complex with diphenyl indole inhibitor VX3 | | Descriptor: | 2,3-diphenyl-1H-indole-7-carboxylic acid, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Jacobs, M.D. | | Deposit date: | 2007-11-27 | | Release date: | 2007-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Docking study yields four novel inhibitors of the protooncogene pim-1 kinase.
J.Med.Chem., 51, 2008
|
|
7KKE
 
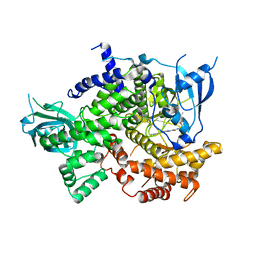 | | Phosphoinositide 3-Kinase gamma bound to a thiazole inhibitor | | Descriptor: | N-[2-(3,3-dimethylbutoxy)ethyl]-N'-{4-methyl-5-[(pyridin-4-yl)ethynyl]-1,3-thiazol-2-yl}urea, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Jacobs, M.D, Griffith, J.P. | | Deposit date: | 2020-10-27 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Novel Series of Potent and Selective Alkynylthiazole-Derived PI3K gamma Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
2OJG
 
 | |
2OJI
 
 | |
2OK1
 
 | |
2OJJ
 
 | |
3I4B
 
 | | Crystal structure of GSK3b in complex with a pyrimidylpyrrole inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-[(1S)-2-hydroxy-1-phenylethyl]-4-[5-methyl-2-(phenylamino)pyrimidin-4-yl]-1H-pyrrole-2-carboxamide | | Authors: | Ter Haar, E. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control.
J.Med.Chem., 52, 2009
|
|
1MGX
 
 | | COAGULATION FACTOR, MG(II), NMR, 7 STRUCTURES (BACKBONE ATOMS ONLY) | | Descriptor: | COAGULATION FACTOR IX | | Authors: | Freedman, S.J, Furie, B.C, Furie, B, Baleja, J.D. | | Deposit date: | 1995-06-21 | | Release date: | 1996-11-08 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Identification of the phospholipid binding site in the vitamin K-dependent blood coagulation protein factor IX.
J.Biol.Chem., 271, 1996
|
|
3CUK
 
 | |
7UN5
 
 | |
7UMQ
 
 | |
1USL
 
 | | Structure Of Mycobacterium tuberculosis Ribose-5-Phosphate Isomerase, RpiB, Rv2465c, Complexed With Phosphate. | | Descriptor: | PHOSPHATE ION, RIBOSE 5-PHOSPHATE ISOMERASE B | | Authors: | Roos, A.K, Andersson, C.E, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2003-11-25 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mycobacterium Tuberculosis Ribose-5-Phosphate Isomerase Has a Known Fold, But a Novel Active Site
J.Mol.Biol., 335, 2004
|
|
2BET
 
 | | Structure of Mycobacterium tuberculosis Ribose-5-Phosphate Isomerase, RpiB, Rv2465c, in complex with 4-phospho-D-erythronate. | | Descriptor: | 4-PHOSPHO-D-ERYTHRONATE, CARBOHYDRATE-PHOSPHATE ISOMERASE | | Authors: | Roos, A.K, Ericsson, D.J, Mowbray, S.L. | | Deposit date: | 2004-11-30 | | Release date: | 2004-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Competitive Inhibitors of Mycobacterium Tuberculosis Ribose-5-Phosphate Isomerase B Reveal New Information About the Reaction Mechanism
J.Biol.Chem., 280, 2005
|
|
2BES
 
 | | Structure of Mycobacterium tuberculosis Ribose-5-Phosphate Isomerase, RpiB, Rv2465c, in complex with 4-phospho-D-erythronohydroxamic acid. | | Descriptor: | 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID, CARBOHYDRATE-PHOSPHATE ISOMERASE | | Authors: | Roos, A.K, Ericsson, D.J, Mowbray, S.L. | | Deposit date: | 2004-11-30 | | Release date: | 2004-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Competitive Inhibitors of Mycobacterium Tuberculosis Ribose-5-Phosphate Isomerase B Reveal New Information About the Reaction Mechanism.
J.Biol.Chem., 280, 2005
|
|
