6K0V
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with tetrasaccharides | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0S
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
1VD5
 
 | | Crystal Structure of Unsaturated Glucuronyl Hydrolase, Responsible for the Degradation of Glycosaminoglycan, from Bacillus sp. GL1 at 1.8 A Resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCINE, ... | | Authors: | Itoh, T, Akao, S, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-03-18 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Unsaturated Glucuronyl Hydrolase, Responsible for the Degradation of Glycosaminoglycan, from Bacillus sp. GL1 at 1.8 A Resolution
J.Biol.Chem., 279, 2004
|
|
5I04
 
 | | Crystal structure of the orphan region of human endoglin/CD105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltose-binding periplasmic protein,Endoglin, TRIETHYLENE GLYCOL, ... | | Authors: | Saito, T, Bokhove, M, de Sanctis, D, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
5ZEQ
 
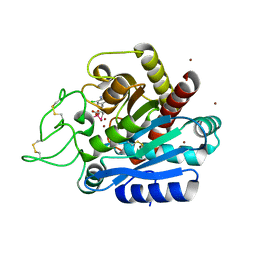 | | Carboxypeptidase B in complex with DD28 | | Descriptor: | (2~{S})-2-[(6-azanyl-5-chloranyl-pyridin-3-yl)methyl]-3-selanyl-propanoic acid, CACODYLATE ION, Carboxypeptidase B, ... | | Authors: | Itoh, T, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the selective inhibition of activated thrombin-activatable fibrinolysis inhibitor (TAFIa) by a selenium-containing inhibitor with chloro-aminopyridine as a basic group
Bioorg. Med. Chem. Lett., 28, 2018
|
|
3AX3
 
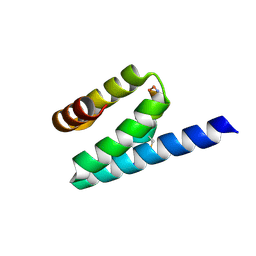 | |
3AX2
 
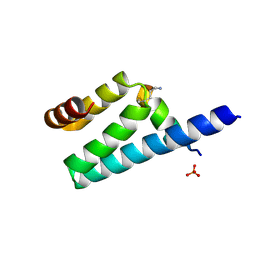 | | Crystal structure of rat TOM20-ALDH presequence complex: a disulfide-tethered complex with a non-optimized, long linker | | Descriptor: | Aldehyde dehydrogenase, mitochondrial, Mitochondrial import receptor subunit TOM20 homolog, ... | | Authors: | Saitoh, T, Maita, Y, Kohda, D. | | Deposit date: | 2011-03-28 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic snapshots of tom20-mitochondrial presequence interactions with disulfide-stabilized peptides.
Biochemistry, 50, 2011
|
|
3AX5
 
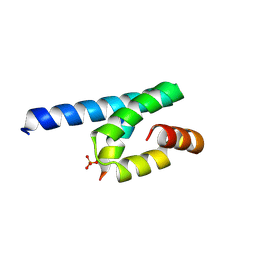 | | Crystal structure of rat TOM20-ALDH presequence complex: A complex (form1) between Tom20 and a disulfide-bridged presequence peptide containing D-Cys and L-Cys at the i and i+3 positions. | | Descriptor: | Aldehyde dehydrogenase, mitochondrial, Mitochondrial import receptor subunit TOM20 homolog, ... | | Authors: | Saitoh, T, Maita, Y, Kohda, D. | | Deposit date: | 2011-03-29 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic snapshots of tom20-mitochondrial presequence interactions with disulfide-stabilized peptides.
Biochemistry, 50, 2011
|
|
2RGK
 
 | | Functional annotation of Escherichia coli yihS-encoded protein | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Uncharacterized sugar isomerase yihS | | Authors: | Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-03 | | Release date: | 2008-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of YihS in complex with D-mannose: structural annotation of Escherichia coli and Salmonella enterica yihS-encoded proteins to an aldose-ketose isomerase
J.Mol.Biol., 377, 2008
|
|
2VV1
 
 | | hPPARgamma Ligand binding domain in complex with 4-HDHA | | Descriptor: | (4S,5E,7Z,10Z,13Z,16Z,19Z)-4-hydroxydocosa-5,7,10,13,16,19-hexaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VSR
 
 | | hPPARgamma Ligand binding domain in complex with 9-(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2AHF
 
 | |
7C7D
 
 | | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3 | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, alpha-1,3-glucanase | | Authors: | Itoh, T, Panti, N, Toyotake, Y, Hayashi, J, Suyotha, W, Yano, S, Wakayama, M, Hibi, T. | | Deposit date: | 2020-05-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7EHP
 
 | | Chitin oligosaccharide binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, chitin oligosaccahride binding protein NagB1 | | Authors: | Itoh, T, Hibi, T, Kimoto, H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural characterization of two solute-binding proteins for N,N' -diacetylchitobiose/ N,N',N'' -triacetylchitotoriose of the gram-positive bacterium, Paenibacillus sp. str. FPU-7.
J Struct Biol X, 5, 2021
|
|
7EHO
 
 | | Chitin oligosaccharide binding protein | | Descriptor: | Chitin oligosaccharide binding protein NagB2, TETRAETHYLENE GLYCOL | | Authors: | Itoh, T, Hibi, T, Kimoto, H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural characterization of two solute-binding proteins for N,N' -diacetylchitobiose/ N,N',N'' -triacetylchitotoriose of the gram-positive bacterium, Paenibacillus sp. str. FPU-7.
J Struct Biol X, 5, 2021
|
|
7EHU
 
 | | Chitin oligosaccharide binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin oligosaccharide binding protein NagB2, DI(HYDROXYETHYL)ETHER | | Authors: | Itoh, T, Hibi, T, Kimoto, H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural characterization of two solute-binding proteins for N,N' -diacetylchitobiose/ N,N',N'' -triacetylchitotoriose of the gram-positive bacterium, Paenibacillus sp. str. FPU-7.
J Struct Biol X, 5, 2021
|
|
7EHQ
 
 | | Chitin oligosaccharide binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin oligosaccharide binding protein NagB2 | | Authors: | Itoh, T, Hibi, T, Kimoto, H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of two solute-binding proteins for N,N' -diacetylchitobiose/ N,N',N'' -triacetylchitotoriose of the gram-positive bacterium, Paenibacillus sp. str. FPU-7.
J Struct Biol X, 5, 2021
|
|
2CV6
 
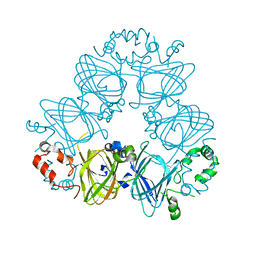 | | Crystal Structure of 8Salpha Globulin, the Major Seed Storage Protein of Mungbean | | Descriptor: | Seed storage protein | | Authors: | Itoh, T, Garcia, R.N, Adachi, M, Maruyama, Y, Tecson-Mendoza, E.M, Mikami, B, Utsumi, S. | | Deposit date: | 2005-05-31 | | Release date: | 2006-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of 8Salpha globulin, the major seed storage protein of mung bean.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2AHG
 
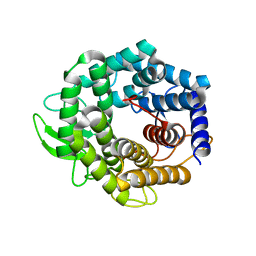 | | Unsaturated glucuronyl hydrolase mutant D88N with dGlcA-GalNAc | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, unsaturated glucuronyl hydrolase | | Authors: | Itoh, T, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2005-07-28 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Unsaturated Glucuronyl Hydrolase Complexed with Substrate: MOLECULAR INSIGHTS INTO ITS CATALYTIC REACTION MECHANISM
J.Biol.Chem., 281, 2006
|
|
2D5J
 
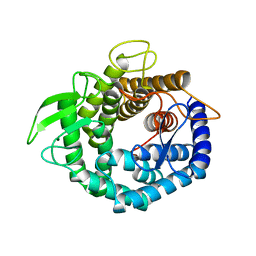 | |
2EVX
 
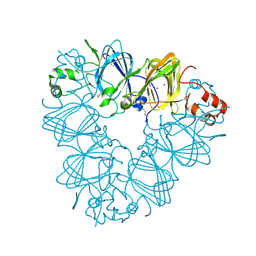 | |
2FUZ
 
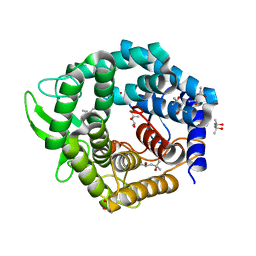 | | UGL hexagonal crystal structure without glycine and DTT molecules | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Unsaturated glucuronyl hydrolase | | Authors: | Itoh, T, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2006-01-28 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate recognition by unsaturated glucuronyl hydrolase from Bacillus sp. GL1
Biochem.Biophys.Res.Commun., 344, 2006
|
|
2FV0
 
 | | UGL_D88N/dGlcA-Glc-Rha-Glc | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-4)-beta-D-glucopyranose-(1-4)-alpha-L-rhamnopyranose-(1-3)-beta-D-glucopyranose, Unsaturated glucuronyl hydrolase | | Authors: | Itoh, T, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2006-01-28 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Substrate recognition by unsaturated glucuronyl hydrolase from Bacillus sp. GL1
Biochem.Biophys.Res.Commun., 344, 2006
|
|
2FV1
 
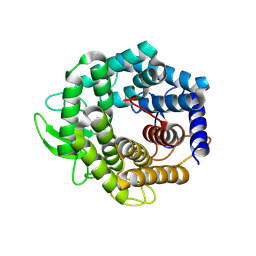 | | UGL_D88N/dGlcA-GlcNAc | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Unsaturated glucuronyl hydrolase | | Authors: | Itoh, T, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2006-01-28 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Substrate recognition by unsaturated glucuronyl hydrolase from Bacillus sp. GL1
Biochem.Biophys.Res.Commun., 344, 2006
|
|
2GH4
 
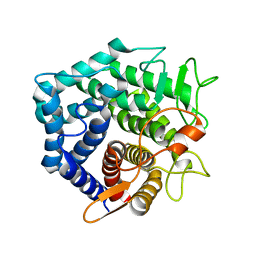 | | YteR/D143N/dGalA-Rha | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, Putative glycosyl hydrolase yteR | | Authors: | Itoh, T, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2006-03-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of unsaturated rhamnogalacturonyl hydrolase complexed with substrate
Biochem.Biophys.Res.Commun., 347, 2006
|
|
