3H4M
 
 | | AAA ATPase domain of the proteasome- activating nucleotidase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Proteasome-activating nucleotidase | | Authors: | Jeffrey, P, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
3H43
 
 | | N-terminal domain of the proteasome-activating nucleotidase of Methanocaldococcus jannaschii | | Descriptor: | Proteasome-activating nucleotidase | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-17 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
3H4P
 
 | | Proteasome 20S core particle from Methanocaldococcus jannaschii | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
8KGC
 
 | |
7CQT
 
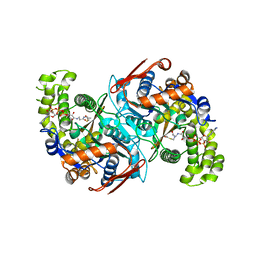 | | Crystal structure of Brassica juncea HMG-CoA synthase 1 mutant - S359A in complex with acetyl-CoA | | Descriptor: | 3-hydroxy-3-methylglutaryl coenzyme A synthase, ACETYL COENZYME *A, ADENOSINE-3'-5'-DIPHOSPHATE | | Authors: | Liao, P, Hu, M, Kong, G.K.W, Hao, Q, Chye, M.L. | | Deposit date: | 2020-08-11 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Overexpression and Inhibition of 3-Hydroxy-3-Methylglutaryl-CoA Synthase Affect Central Metabolic Pathways in Tobacco.
Plant Cell.Physiol., 62, 2021
|
|
7F7W
 
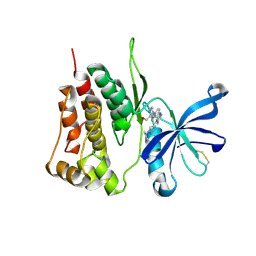 | | JAK2-JH2 | | Descriptor: | 2-((1-(2-fluoro-4-((4-(1-isopropyl-1H-pyrazol-4-yl)-5-methylpyrimidin-2-yl)amino)phenyl)piperidin-4-yl)(methyl)amino)ethan-1-ol, Tyrosine-protein kinase JAK2 | | Authors: | Niu, L. | | Deposit date: | 2021-06-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Preclinical studies of Flonoltinib Maleate, a novel JAK2/FLT3 inhibitor, in treatment of JAK2 V617F -induced myeloproliferative neoplasms.
Blood Cancer J, 12, 2022
|
|
1HTV
 
 | |
8YJO
 
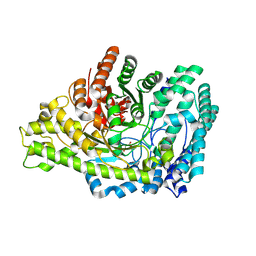 | | Structure of E. coli glycyl radical enzyme PflD with bound malonate | | Descriptor: | MALONATE ION, Probable dehydratase PflD | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
8YJN
 
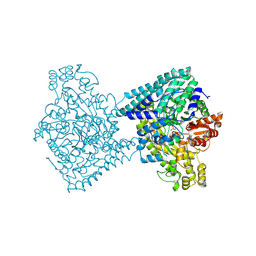 | | Structure of E. coli glycyl radical enzyme YbiW with bound glycerol | | Descriptor: | GLYCEROL, Probable dehydratase YbiW | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
6S1F
 
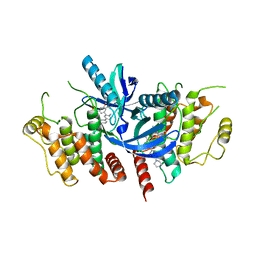 | | Structure of the kinase domain of human RIPK2 in complex with the inhibitor CSLP3 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2, ~{N}-[3-[2-azanyl-5-(4-piperazin-1-ylphenyl)pyridin-3-yl]-5-methoxy-phenyl]methanesulfonamide | | Authors: | Pinkas, D.M, Bufton, J.C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-06-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Design of 3,5-diaryl-2-aminopyridines as receptor-interacting protein kinase 2 (RIPK2) and nucleotide-binding oligomerization domain (NOD) cell signaling inhibitors
To be published
|
|
7JU5
 
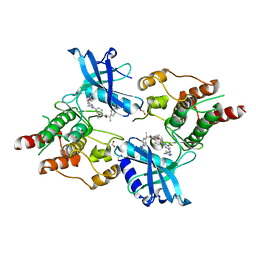 | | Structure of RET protein tyrosine kinase in complex with pralsetinib | | Descriptor: | FORMIC ACID, Pralsetinib, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2020-08-19 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of acquired resistance to selpercatinib and pralsetinib mediated by non-gatekeeper RET mutations.
Ann Oncol, 32, 2021
|
|
7JU6
 
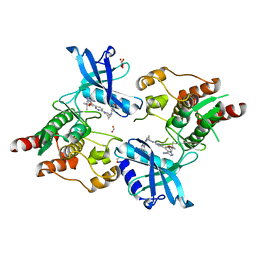 | | Structure of RET protein tyrosine kinase in complex with selpercatinib | | Descriptor: | FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret, Selpercatinib | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2020-08-19 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis of acquired resistance to selpercatinib and pralsetinib mediated by non-gatekeeper RET mutations.
Ann Oncol, 32, 2021
|
|
8XJV
 
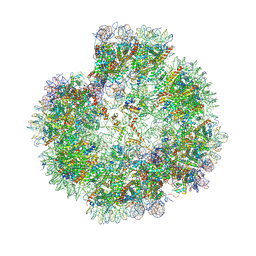 | | Structural basis for the linker histone H5-nucleosome binding and chromatin compaction | | Descriptor: | DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, W.Y, Song, F, Zhu, P. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for linker histone H5-nucleosome binding and chromatin fiber compaction.
Cell Res., 34, 2024
|
|
7FCP
 
 | |
7FCQ
 
 | |
7S8U
 
 | |
3MNR
 
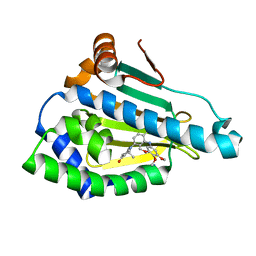 | | Crystal Structure of Benzamide SNX-1321 bound to Hsp90 | | Descriptor: | 2-[(3,4,5-trimethoxyphenyl)amino]-4-(2,6,6-trimethyl-4-oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)benzamide, Heat shock protein HSP 90-alpha | | Authors: | Veal, J.M, Fadden, P, Huang, K.H, Rice, J, Hall, S.E, Haytstead, T.A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Chemoproteomics to Drug Discovery: Identification of a Clinical Candidate Targeting Hsp90.
Chem.Biol., 17, 2010
|
|
4RAP
 
 | | Crystal structure of bacterial iron-containing dodecameric glycosyltransferase TibC from enterotoxigenic E.coli H10407 | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Glycosyltransferase TibC | | Authors: | Yao, Q, Lu, Q, Xu, Y, Shao, F. | | Deposit date: | 2014-09-10 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | A structural mechanism for bacterial autotransporter glycosylation by a dodecameric heptosyltransferase family.
Elife, 3, 2014
|
|
2QXV
 
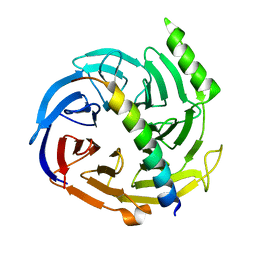 | | Structural basis of EZH2 recognition by EED | | Descriptor: | Embryonic ectoderm development, Enhancer of zeste homolog 2 | | Authors: | Han, Z. | | Deposit date: | 2007-08-13 | | Release date: | 2007-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis of EZH2 recognition by EED
Structure, 15, 2007
|
|
3MYG
 
 | | Aurora A Kinase complexed with SCH 1473759 | | Descriptor: | 2-{ethyl[(5-{[6-methyl-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-yl]amino}isothiazol-3-yl)methyl]amino}-2-methylpropan-1-ol, Serine/threonine-protein kinase 6, TETRAETHYLENE GLYCOL | | Authors: | Hruza, A, Prosis, W, Ramanathan, L. | | Deposit date: | 2010-05-10 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Injectable Inhibitor of Aurora Kinases Based on the Imidazo-[1,2-a]-Pyrazine Core.
ACS Med Chem Lett, 1, 2010
|
|
8ID7
 
 | |
8ID0
 
 | | Crystal structure of PflD bound to 1,5-anhydromannitol-6-phosphate in Streptococcus dysgalactiae subsp. equisimilis | | Descriptor: | [(2R,3S,4R,5R)-3,4,5-tris(oxidanyl)oxan-2-yl]methyl dihydrogen phosphate, formate C-acetyltransferase | | Authors: | Ma, K.L, Zhang, Y. | | Deposit date: | 2023-02-11 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
1I6M
 
 | |
1I6K
 
 | |
6ADL
 
 | |
