2EEP
 
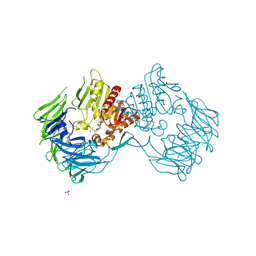 | | Prolyl Tripeptidyl Aminopeptidase Complexed with an Inhibitor | | Descriptor: | Dipeptidyl aminopeptidase IV, putative, SULFATE ION, ... | | Authors: | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2007-02-16 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
6CVH
 
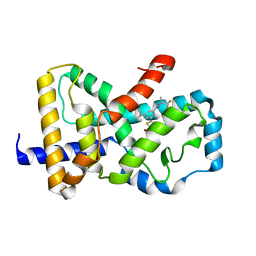 | | Identification and biological evaluation of thiazole-based inverse agonists of RORgt | | Descriptor: | Nuclear receptor ROR-gamma, trans-3-({4-(cyclohexylmethyl)-5-[3-(1-methylcyclopropyl)-5-{[(2R)-1,1,1-trifluoropropan-2-yl]carbamoyl}phenyl]-1,3-thiazole-2-carbonyl}amino)cyclobutane-1-carboxylic acid | | Authors: | Spurlino, J, Milligan, C. | | Deposit date: | 2018-03-28 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Identification and biological evaluation of thiazole-based inverse agonists of ROR gamma t.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6VSW
 
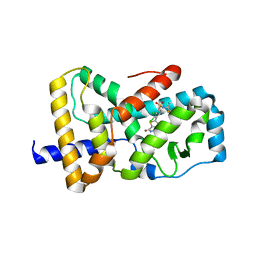 | | Optimization and biological evaluation of thiazole-bis-amide inverse agonists of RORgt | | Descriptor: | 5-(2,3-dichloro-4-{[(2S)-1,1,1-trifluoropropan-2-yl]sulfamoyl}phenyl)-4-(4-fluoropiperidine-1-carbonyl)-N-(2-hydroxy-2-methylpropyl)-1,3-thiazole-2-carboxamide, RAR-related orphan receptor C | | Authors: | Spurlino, J, Milligan, C. | | Deposit date: | 2020-02-12 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Optimization and biological evaluation of thiazole-bis-amide inverse agonists of ROR gamma t.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6QB5
 
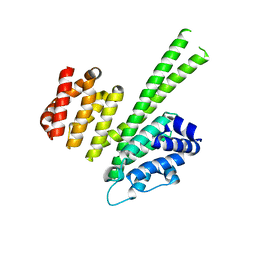 | | Crystal structure of the N-terminal region of human cohesin subunit STAG1 | | Descriptor: | Cohesin subunit SA-1, SODIUM ION | | Authors: | Newman, J.A, Katis, V.L, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-12-20 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | STAG1 vulnerabilities for exploiting cohesin synthetic lethality in STAG2-deficient cancers.
Life Sci Alliance, 3, 2020
|
|
2Z3Z
 
 | | Prolyl tripeptidyl aminopeptidase mutant E636A complexd with an inhibitor | | Descriptor: | Dipeptidyl aminopeptidase IV, SULFATE ION, [(2R)-1-(L-ALANYL-L-ISOLEUCYL)PYRROLIDIN-2-YL]BORONIC ACID | | Authors: | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2007-06-09 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
8AT2
 
 | | Structure of the augmin TIII subcomplex | | Descriptor: | HAUS augmin like complex subunit 4 L homeolog, HAUS augmin-like complex subunit 1, HAUS augmin-like complex subunit 3, ... | | Authors: | Zupa, E, Pfeffer, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | The augmin complex architecture reveals structural insights into microtubule branching.
Nat Commun, 13, 2022
|
|
8AT3
 
 | | Structure of the augmin holocomplex in open conformation | | Descriptor: | HAUS augmin like complex subunit 2 L homeolog, HAUS augmin like complex subunit 4 L homeolog, HAUS augmin like complex subunit 6 L homeolog, ... | | Authors: | Zupa, E, Pfeffer, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (33 Å) | | Cite: | The augmin complex architecture reveals structural insights into microtubule branching.
Nat Commun, 13, 2022
|
|
8AT4
 
 | | Structure of the augmin holocomplex in closed conformation | | Descriptor: | HAUS augmin like complex subunit 2 L homeolog, HAUS augmin like complex subunit 4 L homeolog, HAUS augmin like complex subunit 6 L homeolog, ... | | Authors: | Zupa, E, Pfeffer, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (33 Å) | | Cite: | The augmin complex architecture reveals structural insights into microtubule branching.
Nat Commun, 13, 2022
|
|
2Z3W
 
 | | Prolyl tripeptidyl aminopeptidase mutant E636A | | Descriptor: | Dipeptidyl aminopeptidase IV, GLYCEROL, SULFATE ION | | Authors: | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2007-06-07 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
7BJL
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-142 | | Descriptor: | 14-3-3 protein sigma, 3-[(3~{S})-3-methoxypiperidin-1-yl]-4-nitro-benzaldehyde, Transcription factor p65 | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BJW
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-154 | | Descriptor: | 14-3-3 protein sigma, 4-nitro-3-(4-oxidanylpiperidin-1-yl)benzaldehyde, Transcription factor p65 | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BIQ
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-180 | | Descriptor: | 14-3-3 protein sigma, 4-[[(2~{R})-2-methyl-3,4-dihydro-2~{H}-quinolin-1-yl]sulfonyl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BJF
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-069 | | Descriptor: | (5-methyl-2-nitro-phenyl) cyclobutanecarboxylate, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BKH
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-153 | | Descriptor: | 14-3-3 protein sigma, 4-nitro-3-[(3S)-3-oxidanylpiperidin-1-yl]benzaldehyde, Transcription factor p65 | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BI3
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-179 | | Descriptor: | 14-3-3 protein sigma, 4-[(6-methoxy-3,4-dihydro-2~{H}-quinolin-1-yl)sulfonyl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BIY
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-175 | | Descriptor: | 1-(4-methanoylphenyl)carbonylpiperidine-4-carbonitrile, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BIW
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-187 | | Descriptor: | 14-3-3 protein sigma, 4-(3,4-dihydro-2~{H}-quinoxalin-1-ylsulfonyl)benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BJB
 
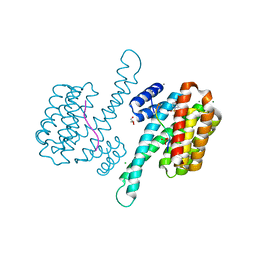 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-044 | | Descriptor: | 14-3-3 protein sigma, 4-(4-methylphenyl)sulfonylmorpholine, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
6R7O
 
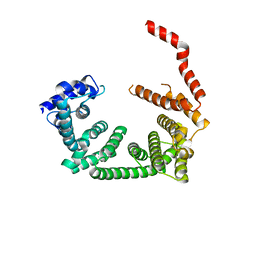 | | Crystal structure of the central region of human cohesin subunit STAG1 | | Descriptor: | Cohesin subunit SA-1 | | Authors: | Newman, J.A, katis, V.L, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-03-29 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | STAG1 vulnerabilities for exploiting cohesin synthetic lethality in STAG2-deficient cancers.
Life Sci Alliance, 3, 2020
|
|
6RRK
 
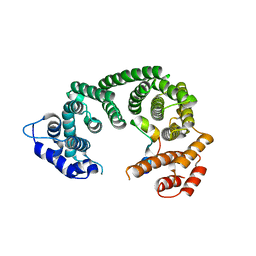 | | Crystal structure of the central region of human cohesin subunit STAG1 in complex with RAD21 peptide | | Descriptor: | Cohesin subunit SA-1, Double-strand-break repair protein rad21 homolog | | Authors: | Newman, J.A, katis, V.L, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-20 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | STAG1 vulnerabilities for exploiting cohesin synthetic lethality in STAG2-deficient cancers.
Life Sci Alliance, 3, 2020
|
|
6RRC
 
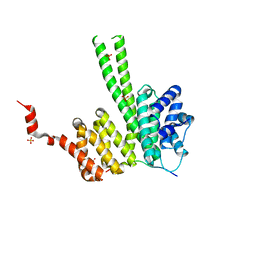 | | Crystal structure of the N-terminal region of human cohesin subunit STAG1 in complex with RAD21 peptide | | Descriptor: | Cohesin subunit SA-1, Double-strand-break repair protein rad21 homolog, SULFATE ION | | Authors: | Newman, J.A, Katis, V.L, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-17 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | STAG1 vulnerabilities for exploiting cohesin synthetic lethality in STAG2-deficient cancers.
Life Sci Alliance, 3, 2020
|
|
6YOY
 
 | |
6YP2
 
 | |
6YP3
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-028 | | Descriptor: | 1-(4-methylphenyl)-1,2,4-triazole, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2020-04-15 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Stabilizers of Protein-Protein Interactions through Imine-Based Tethering.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6YP8
 
 | |
