8JES
 
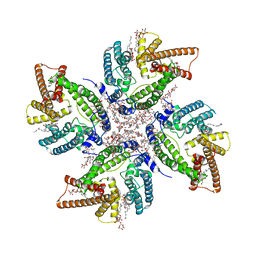 | | Cryo-EM structure of DltB homo-tetramer | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, DIACYL GLYCEROL, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Zhang, P, Liu, Z. | | Deposit date: | 2023-05-16 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural insights into the transporting and catalyzing mechanism of DltB in LTA D-alanylation.
Nat Commun, 15, 2024
|
|
8JF2
 
 | | Cryo-EM structure of tetrameric DltB/DltC complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, D-alanyl carrier protein, DIACYL GLYCEROL, ... | | Authors: | Zhang, P, Liu, Z. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the transporting and catalyzing mechanism of DltB in LTA D-alanylation.
Nat Commun, 15, 2024
|
|
2F5K
 
 | |
2VKZ
 
 | | Structure of the cerulenin-inhibited fungal fatty acid synthase type I multienzyme complex | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, FATTY ACID SYNTHASE SUBUNIT ALPHA, ... | | Authors: | Johansson, P, Wiltschi, B, Kumari, P, Kessler, B, Vonrhein, C, Vonck, J, Oesterhelt, D, Grininger, M. | | Deposit date: | 2008-01-07 | | Release date: | 2008-08-12 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Inhibition of the Fungal Fatty Acid Synthase Type I Multienzyme Complex.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
8KGC
 
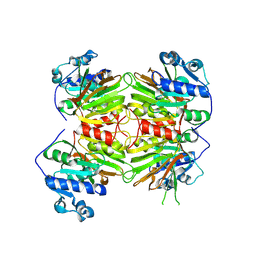 | |
8P5O
 
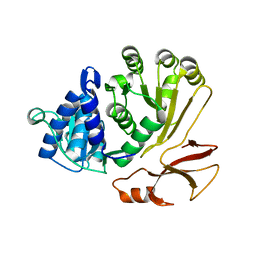 | | Proline activating adenylation domain of gramicidin S synthetase 2 - GrsB1-Acore | | Descriptor: | Gramicidin S synthase 2 | | Authors: | Stephan, P, Basquin, J, Caputi, L, O'Connor, S.E, Kries, H. | | Deposit date: | 2023-05-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Directed Evolution of Piperazic Acid Incorporation by a Nonribosomal Peptide Synthetase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3F6H
 
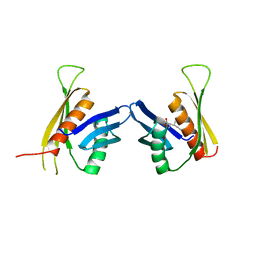 | | Crystal structure of the regulatory domain of LiCMS in complexed with isoleucine - type III | | Descriptor: | Alpha-isopropylmalate synthase, ISOLEUCINE, ZINC ION | | Authors: | Zhang, P, Ma, J, Zhao, G, Ding, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of the inhibitor selectivity and insights into the feedback inhibition mechanism of citramalate synthase from Leptospira interrogans
Biochem.J., 421, 2009
|
|
3F6G
 
 | | Crystal structure of the regulatory domain of LiCMS in complexed with isoleucine - type II | | Descriptor: | Alpha-isopropylmalate synthase, ISOLEUCINE, SULFATE ION, ... | | Authors: | Zhang, P, Ma, J, Zhao, G, Ding, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of the inhibitor selectivity and insights into the feedback inhibition mechanism of citramalate synthase from Leptospira interrogans
Biochem.J., 421, 2009
|
|
3EPC
 
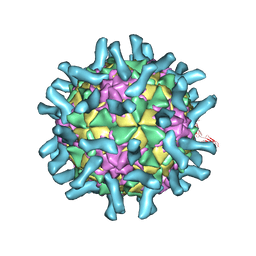 | | CryoEM structure of poliovirus receptor bound to poliovirus type 1 | | Descriptor: | MYRISTIC ACID, Poliovirus receptor, Protein VP1, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EPF
 
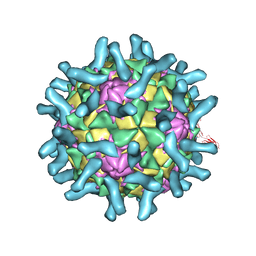 | | CryoEM structure of poliovirus receptor bound to poliovirus type 2 | | Descriptor: | 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE, MYRISTIC ACID, Poliovirus receptor, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EPD
 
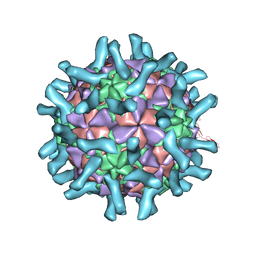 | | CryoEM structure of poliovirus receptor bound to poliovirus type 3 | | Descriptor: | MYRISTIC ACID, Poliovirus Type3 peptide, Poliovirus receptor, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3P5N
 
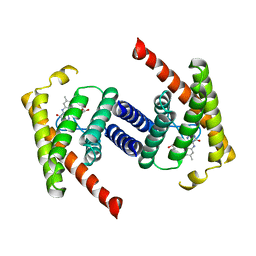 | |
7AI3
 
 | |
7AI2
 
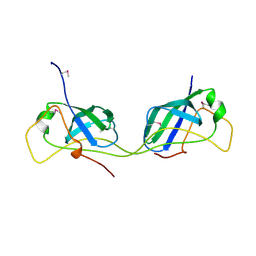 | |
1DRO
 
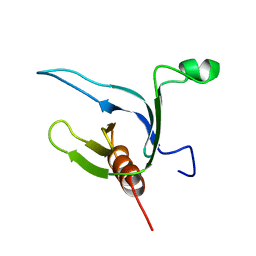 | | NMR STRUCTURE OF THE CYTOSKELETON/SIGNAL TRANSDUCTION PROTEIN | | Descriptor: | BETA-SPECTRIN | | Authors: | Zhang, P, Talluri, S, Deng, H, Branton, D, Wagner, G. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the pleckstrin homology domain of Drosophila beta-spectrin.
Structure, 3, 1995
|
|
7E5Q
 
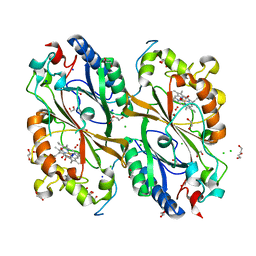 | | Crystal Structure of Dye Decolorizing peroxidase from Bacillus subtilis at acidic pH | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Dhankhar, P, Dalal, V, Kumar, P. | | Deposit date: | 2021-02-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights at acidic pH of dye-decolorizing peroxidase from Bacillus subtilis.
Proteins, 2022
|
|
2WAT
 
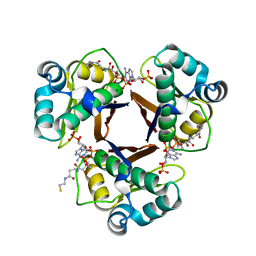 | | Structure of the fungal type I FAS PPT domain in complex with CoA | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE, CHLORIDE ION, COENZYME A, ... | | Authors: | Johansson, P, Mulincacci, B, Koestler, C, Grininger, M. | | Deposit date: | 2009-02-15 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multimeric Options for the Auto-Activation of the Saccharomyces Cerevisiae Fas Type I Megasynthase.
Structure, 17, 2009
|
|
2WAS
 
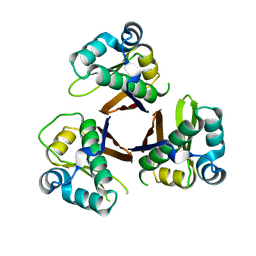 | |
1UN1
 
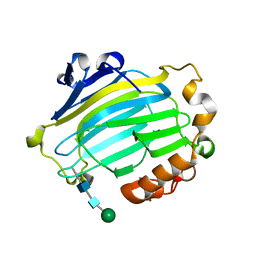 | | Xyloglucan endotransglycosylase native structure. | | Descriptor: | GOLD ION, XYLOGLUCAN ENDOTRANSGLYCOSYLASE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Johansson, P, Brumer, H, Kallas, A, Henriksson, H, Denman, S, Teeri, T.T, Jones, T.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-03-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of a Poplar Xyloglucan Endotransglycosylase Reveal Details of Transglycosylation Acceptor Binding
Plant Cell, 16, 2004
|
|
7FHA
 
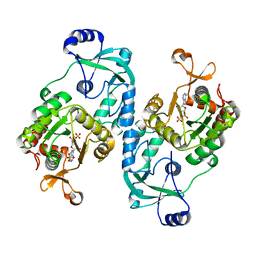 | | Crystal structure of the ATP sulfurylase domain of human PAPSS2 in complex with APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthase 2, POTASSIUM ION, ... | | Authors: | Zhang, P, Zhang, L, Zhang, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the substrate recognition mechanism of ATP-sulfurylase domain of human PAPS synthase 2.
Biochem.Biophys.Res.Commun., 586, 2022
|
|
7FH3
 
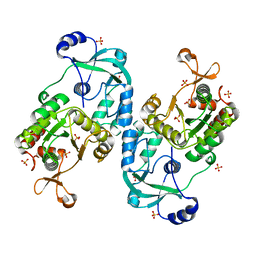 | | Crystal structure of the ATP sulfurylase domain of human PAPSS2 | | Descriptor: | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthase 2, SULFATE ION, beta-D-glucopyranose | | Authors: | Zhang, P, Zhang, L, Zhang, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the substrate recognition mechanism of ATP-sulfurylase domain of human PAPS synthase 2.
Biochem.Biophys.Res.Commun., 586, 2022
|
|
4AJD
 
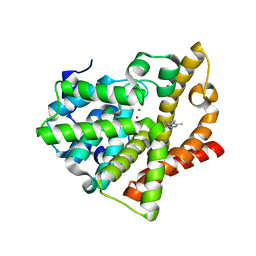 | | Identification and structural characterization of PDE10 fragment inhibitors | | Descriptor: | 2-ETHYL-4-METHYL-PHTHALAZIN-1-ONE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Johansson, P, Albert, J.S, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Geschwindner, S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and Structural Characterization of Pde10 Fragment Inhibitors
To be Published
|
|
4AJF
 
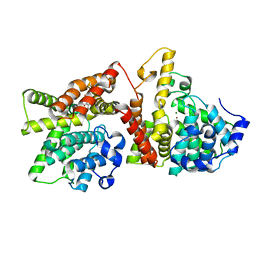 | | Identification and structural characterization of PDE10 fragment inhibitors | | Descriptor: | CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, MAGNESIUM ION, ... | | Authors: | Johansson, P, Albert, J.S, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Geschwindner, S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Structural Characterization of Pde10 Fragment Inhibitors
To be Published
|
|
1XZN
 
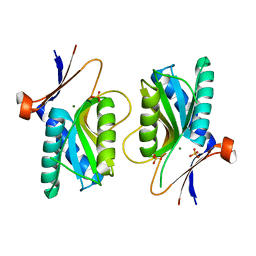 | | PYRR, THE REGULATOR OF THE PYRIMIDINE BIOSYNTHETIC OPERON IN BACILLUS CALDOLYTICUS, sulfate-bound form | | Descriptor: | MAGNESIUM ION, PyrR bifunctional protein, SULFATE ION | | Authors: | Chander, P, Halbig, K.M, Miller, J.K, Fields, C.J, Bonner, H.K, Grabner, G.K, Switzer, R.L, Smith, J.L. | | Deposit date: | 2004-11-12 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of the Nucleotide Complex of PyrR, the pyr Attenuation Protein from Bacillus caldolyticus, Suggests Dual Regulation by Pyrimidine and Purine Nucleotides.
J.Bacteriol., 187, 2005
|
|
1JDG
 
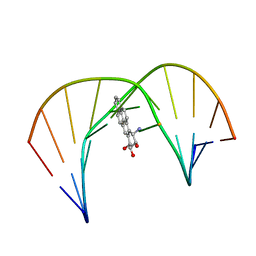 | | Solution Structure of a Trans-Opened (10S)-dA Adduct of (+)-(7S,8R,9S,10R)-7,8-Dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a fully Complementary DNA Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*GP*TP*GP*AP*CP*CP*G)-3', 5'-D(*CP*GP*GP*TP*CP*(BPA)AP*CP*GP*AP*GP*G)-3', 7S,8R,9R-TRIHYDROXY-7,8,9,10-TETRAHYDRO BENZO[A]PYRENE | | Authors: | Pradhan, P, Tirumala, S, Liu, X, Sayer, J.M, Jerina, D.M, Yeh, H.J.C. | | Deposit date: | 2001-06-13 | | Release date: | 2001-07-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a trans-opened (10S)-dA adduct of (+)-(7S,8R,9S,10R)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a fully complementary DNA duplex: evidence for a major syn conformation.
Biochemistry, 40, 2001
|
|
