3OZB
 
 | | Crystal Structure of 5'-methylthioinosine phosphorylase from Psedomonas aeruginosa in complex with hypoxanthine | | Descriptor: | HYPOXANTHINE, Methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Ho, M, Guan, R, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-09-24 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Methylthioinosine phosphorylase from Pseudomonas aeruginosa. Structure and annotation of a novel enzyme in quorum sensing.
Biochemistry, 50, 2011
|
|
8EVJ
 
 | | CX3CR1 nucleosome bound PU.1 and C/EBPa | | Descriptor: | DNA (167-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Guan, R, Bai, Y, Lian, T. | | Deposit date: | 2022-10-20 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural mechanism of synergistic targeting of the CX3CR1 nucleosome by PU.1 and C/EBP alpha.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7U0G
 
 | | structure of LIN28b nucleosome bound 3 OCT4 | | Descriptor: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Lian, T, Guan, R, Bai, Y. | | Deposit date: | 2022-02-18 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
7U0I
 
 | | Structure of LIN28b nucleosome bound 2 OCT4 | | Descriptor: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Tengfei, L, Guan, R, Bai, Y. | | Deposit date: | 2022-02-18 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
7U0J
 
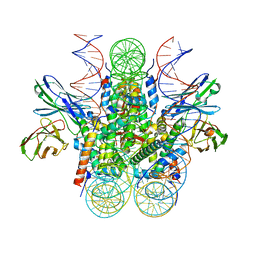 | | Structure of 162bp LIN28b nucleosome | | Descriptor: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Lian, T, Guan, R, Bai, Y. | | Deposit date: | 2022-02-18 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
4XOH
 
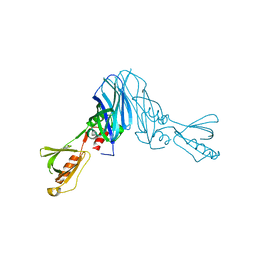 | | Mechanistic insights into anchorage of the contractile ring from yeast to humans | | Descriptor: | Division mal foutue 1 protein | | Authors: | Chen, Z, Wu, J.-Q, Wang, J, Guan, R, Sun, L, Lee, I.-J, Liu, Y, Chen, M. | | Deposit date: | 2015-01-16 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Mechanistic insights into the anchorage of the contractile ring by anillin and mid1
Dev.Cell, 33, 2015
|
|
4XOI
 
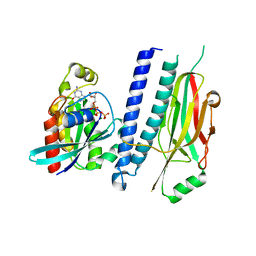 | | Structure of hsAnillin bound with RhoA(Q63L) at 2.1 Angstroms resolution | | Descriptor: | Actin-binding protein anillin, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sun, L, Guan, R, Lee, I.-J, Liu, Y, Chen, M, Wang, J, Wu, J, Chen, Z. | | Deposit date: | 2015-01-16 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Mechanistic insights into the anchorage of the contractile ring by anillin and mid1
Dev.Cell, 33, 2015
|
|
4RJV
 
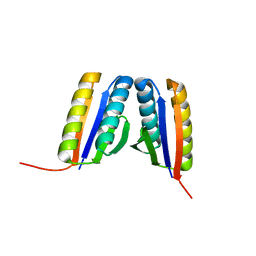 | | Crystal Structure of a De Novo Designed Ferredoxin Fold, Northeast Structural Genomics Consortium (NESG) Target OR461 | | Descriptor: | OR461 | | Authors: | O'Connell, P.T, Lin, Y.-R, Guan, R, Koga, N, Koga, R, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-10-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR461
To be published
|
|
2UZT
 
 | | PKA structures of AKT, indazole-pyridine inhibitors | | Descriptor: | (2S)-1-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}-3-PHENYLPROPAN-2-AMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Zhu, G.D, Gandhi, V.B, Gong, J, Thomas, S, Woods, K.W, Song, X, Li, T, Diebold, R.B, Luo, Y, Liu, X, Guan, R, Klinghofer, V, Johnson, E.F, Bouska, J, Olson, A, Marsh, K.C, Stoll, V.S, Mamo, M, Polakowski, J, Campbell, T.J, Penning, T.D, Li, Q, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Syntheses of Potent, Selective, and Orally Bioavailable Indazole-Pyridine Series of Protein Kinase B/Akt Inhibitors with Reduced Hypotension.
J.Med.Chem., 50, 2007
|
|
2UZU
 
 | | PKA structures of indazole-pyridine series of AKT inhibitors | | Descriptor: | (2S)-1-(1H-INDOL-3-YL)-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Zhu, G.D, Gandhi, V.B, Gong, J, Thomas, S, Woods, K.W, Song, X, Li, T, Diebold, R.B, Luo, Y, Liu, X, Guan, R, Klinghofer, V, Johnson, E.F, Bouska, J, Olson, A, Marsh, K.C, Stoll, V.S, Mamo, M, Polakowski, J, Campbell, T.J, Penning, T.D, Li, Q, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Syntheses of Potent, Selective, and Orally Bioavailable Indazole-Pyridine Series of Protein Kinase B/Akt Inhibitors with Reduced Hypotension.
J.Med.Chem., 50, 2007
|
|
2UZV
 
 | | PKA structures of indazole-pyridine series of AKT inhibitors | | Descriptor: | (2S)-1-[3-(CYCLOHEXYLMETHOXY)PHENYL]-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA,, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Zhu, G.D, Gandhi, V.B, Gong, J, Thomas, S, Woods, K.W, Song, X, Li, T, Diebold, R.B, Luo, Y, Liu, X, Guan, R, Klinghofer, V, Johnson, E.F, Bouska, J, Olson, A, Marsh, K.C, Stoll, V.S, Mamo, M, Polakowski, J, Campbell, T.J, Penning, T.D, Li, Q, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Syntheses of Potent, Selective, and Orally Bioavailable Indazole-Pyridine Series of Protein Kinase B/Akt Inhibitors with Reduced Hypotension.
J.Med.Chem., 50, 2007
|
|
2UZW
 
 | | PKA structures of indazole-pyridine series of AKT inhibitors | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.] PYRAZOLE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Zhu, G.D, Gandhi, V.B, Gong, J, Thomas, S, Woods, K.W, Song, X, Li, T, Diebold, R.B, Luo, Y, Liu, X, Guan, R, Klinghofer, V, Johnson, E.F, Bouska, J, Olson, A, Marsh, K.C, Stoll, V.S, Mamo, M, Polakowski, J, Campbell, T.J, Penning, T.D, Li, Q, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Syntheses of Potent, Selective, and Orally Bioavailable Indazole-Pyridine Series of Protein Kinase B/Akt Inhibitors with Reduced Hypotension.
J.Med.Chem., 50, 2007
|
|
5V3S
 
 | |
2APW
 
 | | Crystal Structure of the G17E/A52V/S54N/K66E/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APV
 
 | | Crystal Structure of the G17E/A52V/S54N/Q72H/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2AQ1
 
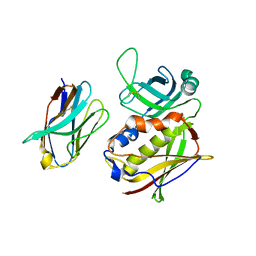 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 mutant | | Descriptor: | Enterotoxin type C-3, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APF
 
 | | Crystal Structure of the A52V/S54N/K66E variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APT
 
 | | Crystal Structure of the G17E/S54N/K66E/Q72H/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2AQ3
 
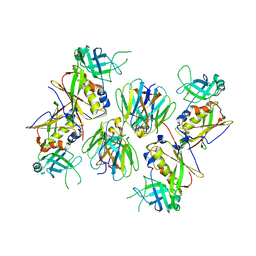 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 | | Descriptor: | Enterotoxin type C-3, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APB
 
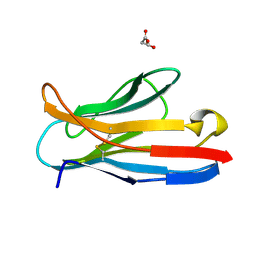 | | Crystal Structure of the S54N variant of murine T cell receptor Vbeta 8.2 domain | | Descriptor: | MALONIC ACID, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APX
 
 | | Crystal Structure of the G17E/A52V/S54N/K66E/Q72H/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2AQ2
 
 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 mutant | | Descriptor: | Enterotoxin type C-3, SODIUM ION, SULFATE ION, ... | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
1U58
 
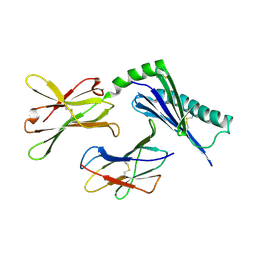 | | Crystal structure of the murine cytomegalovirus MHC-I homolog m144 | | Descriptor: | MHC-I homolog m144, beta-2-microglobulin | | Authors: | Natarajan, K, Hicks, A, Robinson, H, Guan, R, Margulies, D.H. | | Deposit date: | 2004-07-27 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the murine cytomegalovirus MHC-I homolog m144.
J.Mol.Biol., 358, 2006
|
|
2G9H
 
 | | Crystal Structure of Staphylococcal Enterotoxin I (SEI) in Complex with a Human MHC class II Molecule | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HLA class II histocompatibility antigen, ... | | Authors: | Fernandez, M.M, Guan, R, Malchiodi, E.L, Mariuzza, R.A. | | Deposit date: | 2006-03-06 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of staphylococcal enterotoxin I (SEI) in complex with a human major histocompatibility complex class II molecule.
J.Biol.Chem., 281, 2006
|
|
1P4L
 
 | | Crystal structure of NK receptor Ly49C mutant with its MHC class I ligand H-2Kb | | Descriptor: | Beta-2-microglobulin, LY49-C, MHC CLASS I H-2KB HEAVY CHAIN, ... | | Authors: | Dam, J, Guan, R, Natarajan, K, Dimasi, N, Mariuzza, R.A. | | Deposit date: | 2003-04-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Variable MHC class I engagement by Ly49 natural killer cell receptors demonstrated by the crystal structure of Ly49C bound to H-2K(b).
Nat.Immunol., 4, 2003
|
|
