1UOQ
 
 | |
1UOO
 
 | |
1O6G
 
 | |
1O6F
 
 | |
1H2Z
 
 | |
1H2Y
 
 | |
5A9G
 
 | | Manganese Superoxide Dismutase from Sphingobacterium sp. T2 | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Rashid, G.M.M, Taylor, C.R, Liu, Y, Zhang, X, Rea, D, Fulop, V, Bugg, T.D.H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification of Manganese Superoxide Dismutase from Sphingobacterium Sp. T2 as a Novel Bacterial Enzyme for Lignin Oxidation.
Acs Chem.Biol., 10, 2015
|
|
6QSI
 
 | |
5FW4
 
 | | Structure of Thermobifida fusca DyP-type Peroxidase and Activity towards Kraft Lignin and Lignin Model Compounds | | Descriptor: | DYE-DECOLORIZING PEROXIDASE TFU_3078, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rahmanpour, R, Rea, D, Jamshidi, S, Fulop, V, Bugg, T.D.H. | | Deposit date: | 2016-02-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Thermobifida Fusca Dyp-Type Peroxidase and Activity Towards Kraft Lignin and Lignin Model Compounds.
Arch.Biochem.Biophys., 594, 2016
|
|
6GSB
 
 | | Sphingobacterium sp. T2 manganese superoxide dismutase catalyses the oxidative demethylation of polymeric lignin via generation of hydroxyl radical | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Rashid, G.M, Zhang, X, Wilkinson, R.C, Fulop, V, Cottyn, B, Baumberger, S, Bugg, D.H. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Sphingobacterium sp. T2 Manganese Superoxide Dismutase Catalyzes the Oxidative Demethylation of Polymeric Lignin via Generation of Hydroxyl Radical.
ACS Chem. Biol., 13, 2018
|
|
6GSC
 
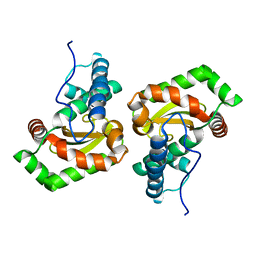 | | Sphingobacterium sp. T2 manganese superoxide dismutase catalyses the oxidative demethylation of polymeric lignin via generation of hydroxyl radical | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Rashid, G.M.M, Zhang, X, Wilkinson, R.C, Fulop, V, Cottyn, B, Baumberger, S, Bugg, T.D.H. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Sphingobacterium sp. T2 Manganese Superoxide Dismutase Catalyzes the Oxidative Demethylation of Polymeric Lignin via Generation of Hydroxyl Radical.
ACS Chem. Biol., 13, 2018
|
|
7KY1
 
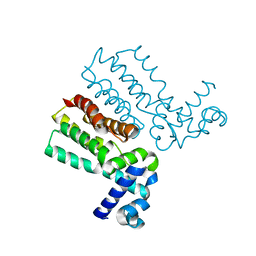 | |
2C1U
 
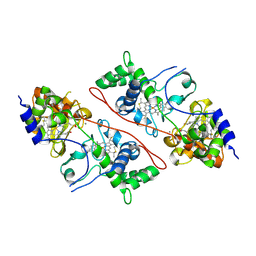 | |
2C1V
 
 | |
2XDW
 
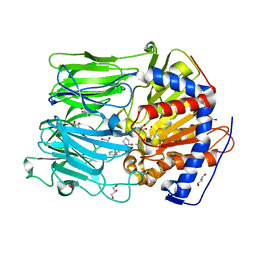 | | Inhibition of Prolyl Oligopeptidase with a Synthetic Unnatural Dipeptide | | Descriptor: | GLYCEROL, Prolyl endopeptidase, SYNTHETIC PEPTIDE PHQ-PRO-YCP, ... | | Authors: | Racys, D.T, Rea, D, Fulop, V, Wills, M. | | Deposit date: | 2010-05-09 | | Release date: | 2010-06-23 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inhibition of Prolyl Oligopeptidase with a Synthetic Unnatural Dipeptide
Bioorg.Med.Chem., 18, 2010
|
|
7ORX
 
 | |
1AOF
 
 | | CYTOCHROME CD1 NITRITE REDUCTASE, REDUCED FORM | | Descriptor: | HEME D, NITRITE REDUCTASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Williams, P.A, Fulop, V. | | Deposit date: | 1997-07-02 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Haem-ligand switching during catalysis in crystals of a nitrogen-cycle enzyme.
Nature, 389, 1997
|
|
1AOM
 
 | |
2VWT
 
 | | Crystal structure of YfaU, a metal ion dependent class II aldolase from Escherichia coli K12 - Mg-pyruvate product complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rea, D, Rakus, J.F, Gerlt, J.A, Fulop, V, Bugg, T.D.H, Roper, D.I. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure and Functional Assignment of Yfau, a Metal Ion Dependent Class II Aldolase from Escherichia Coli K12.
Biochemistry, 47, 2008
|
|
2VWS
 
 | | Crystal structure of YfaU, a metal ion dependent class II aldolase from Escherichia coli K12 | | Descriptor: | GLYCEROL, PHOSPHATE ION, YFAU, ... | | Authors: | Rea, D, Rakus, J.F, Gerlt, J.A, Fulop, V, Bugg, T.D.H, Roper, D.I. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal Structure and Functional Assignment of Yfau, a Metal Ion Dependent Class II Aldolase from Escherichia Coli K12.
Biochemistry, 47, 2008
|
|
1IPS
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (MANGANESE COMPLEX) | | Descriptor: | ISOPENICILLIN N SYNTHASE, MANGANESE (II) ION | | Authors: | Roach, P.L, Clifton, I.J, Fulop, V, Harlos, K, Barton, G.J, Hajdu, J, Andersson, I, Schofield, C.J, Baldwin, J.E. | | Deposit date: | 1997-03-21 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of isopenicillin N synthase is the first from a new structural family of enzymes.
Nature, 375, 1995
|
|
1UQ4
 
 | | RICIN A-CHAIN (RECOMBINANT) R213D MUTANT | | Descriptor: | RICIN, SULFATE ION | | Authors: | Marsden, C.J, Fulop, V. | | Deposit date: | 2003-10-15 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Effect of Mutations Surrounding and within the Active Site on the Catalytic Activity of Ricin a Chain
Eur.J.Biochem., 271, 2004
|
|
1UQ5
 
 | | RICIN A-CHAIN (RECOMBINANT) N122A MUTANT | | Descriptor: | ACETATE ION, RICIN, SULFATE ION | | Authors: | Marsden, C.J, Fulop, V. | | Deposit date: | 2003-10-15 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Effect of Mutations Surrounding and within the Active Site on the Catalytic Activity of Ricin a Chain
Eur.J.Biochem., 271, 2004
|
|
1AOQ
 
 | |
2VC4
 
 | | Ricin A-Chain (Recombinant) E177D Mutant | | Descriptor: | GLYCEROL, RICIN A CHAIN, SULFATE ION | | Authors: | Marsden, C.J, Fulop, V. | | Deposit date: | 2007-09-18 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The Isolation and Characterisation of Temperature-Dependent Ricin a Chain Molecules in Saccharomyces Cerevisiae
FEBS J., 274, 2007
|
|
