9GRZ
 
 | |
9GS3
 
 | |
9GS7
 
 | |
9GS5
 
 | |
8BXG
 
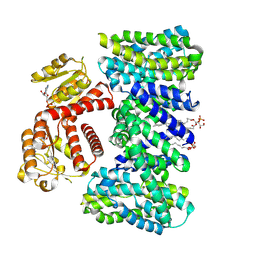 | | Structure of the K/H exchanger KefC. | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ADENOSINE MONOPHOSPHATE, Glutathione-regulated potassium-efflux system protein KefC, ... | | Authors: | Gulati, A, Drew, D. | | Deposit date: | 2022-12-08 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure and mechanism of the K + /H + exchanger KefC.
Nat Commun, 15, 2024
|
|
8BY2
 
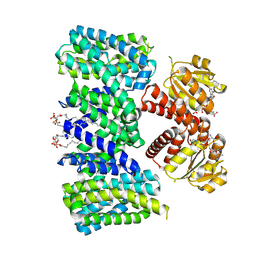 | | Structure of the K+/H+ exchanger KefC with GSH. | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ADENOSINE MONOPHOSPHATE, GLUTATHIONE, ... | | Authors: | Gulati, A, Drew, D. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-27 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structure and mechanism of the K + /H + exchanger KefC.
Nat Commun, 15, 2024
|
|
6I1Z
 
 | |
6I1R
 
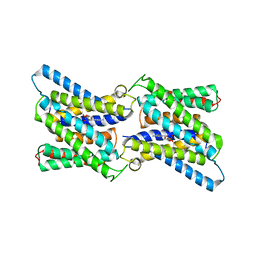 | | Crystal structure of CMP bound CST in an outward facing conformation | | Descriptor: | CMP-sialic acid transporter 1, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Nji, E, Gulati, A, Qureshi, A.A, Drew, D. | | Deposit date: | 2018-10-30 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the delivery of activated sialic acid into Golgi for sialyation.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6RW3
 
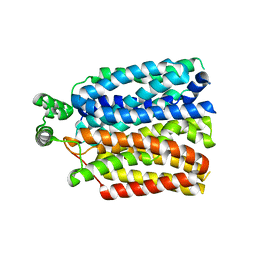 | | The molecular basis for sugar import in malaria parasites. | | Descriptor: | Hexose transporter 1, alpha-D-glucopyranose, beta-D-glucopyranose | | Authors: | Qureshi, A, Matsuoka, R, Brock, J, Drew, D. | | Deposit date: | 2019-06-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The molecular basis for sugar import in malaria parasites.
Nature, 578, 2020
|
|
2XUT
 
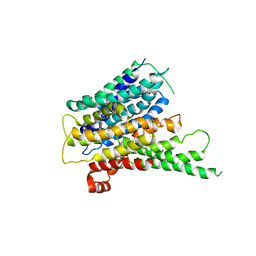 | | Crystal structure of a proton dependent oligopeptide (POT) family transporter. | | Descriptor: | PROTON/PEPTIDE SYMPORTER FAMILY PROTEIN | | Authors: | Newstead, S, Drew, D, Cameron, A.D, Postis, V.L, Xia, X, Fowler, P.W, Ingram, J.C, Carpenter, E.P, Sansom, M.S.P, McPherson, M.J, Baldwin, S.A, Iwata, S. | | Deposit date: | 2010-10-21 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structure of a Prokaryotic Homologue of the Mammalian Oligopeptide-Proton Symporters, Pept1 and Pept2.
Embo J., 30, 2011
|
|
4YB9
 
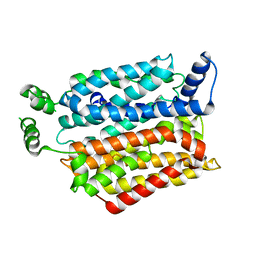 | | Crystal structure of the Bovine Fructose transporter GLUT5 in an open inward-facing conformation | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 5 | | Authors: | Verdon, G, Kang, H.J, Iwata, S, Drew, D. | | Deposit date: | 2015-02-18 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of the mammalian fructose transporter GLUT5.
Nature, 526, 2015
|
|
5BZ3
 
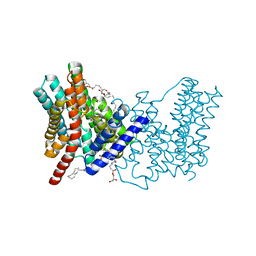 | | CRYSTAL STRUCTURE OF SODIUM PROTON ANTIPORTER NAPA IN OUTWARD-FACING CONFORMATION. | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL (7Z)-TETRADEC-7-ENOATE, Na(+)/H(+) antiporter | | Authors: | Coincon, M, Uzdavinys, P, Emmanuel, N, Cameron, A, Drew, D. | | Deposit date: | 2015-06-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures reveal the molecular basis of ion translocation in sodium/proton antiporters.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5BZ2
 
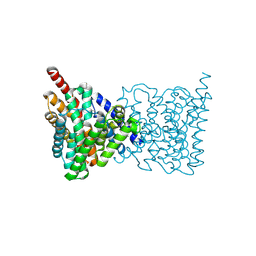 | |
4APS
 
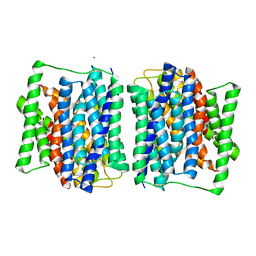 | | Crystal structure of a POT family peptide transporter in an inward open conformation. | | Descriptor: | CADMIUM ION, DI-OR TRIPEPTIDE H+ SYMPORTER | | Authors: | Solcan, N, Kwok, J, Fowler, P.W, Cameron, A.D, Drew, D, Iwata, S, Newstead, S. | | Deposit date: | 2012-04-05 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Alternating Access Mechanism in the Pot Family of Oligopeptide Transporters.
Embo J., 31, 2012
|
|
3ZUY
 
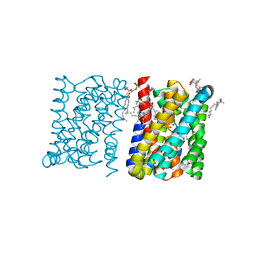 | | Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Hu, N.-J, Iwata, S, Cameron, A.D, Drew, D. | | Deposit date: | 2011-07-21 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Bacterial Homologue of the Bile Acid Sodium Symporter Asbt.
Nature, 478, 2011
|
|
3ZUX
 
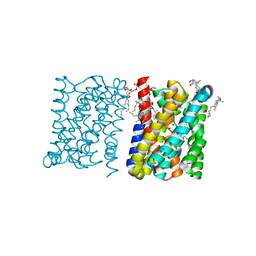 | | Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, MERCURY (II) ION, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Hu, N.-J, Iwata, S, Cameron, A.D, Drew, D. | | Deposit date: | 2011-07-21 | | Release date: | 2011-10-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Bacterial Homologue of the Bile Acid Sodium Symporter Asbt.
Nature, 478, 2011
|
|
4BWZ
 
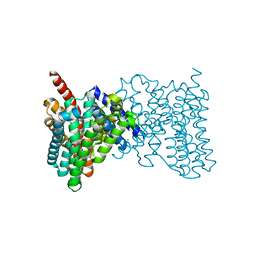 | |
1CGL
 
 | | Structure of the catalytic domain of fibroblast collagenase complexed with an inhibitor | | Descriptor: | CALCIUM ION, FIBROBLAST COLLAGENASE, N-[(1S)-3-{[(benzyloxy)carbonyl]amino}-1-carboxypropyl]-L-leucyl-N-(2-morpholin-4-ylethyl)-L-phenylalaninamide, ... | | Authors: | Lovejoy, B, Cleasby, A, Hassell, A.M, Longley, K, Luther, M.A, Weigl, D, Mcgeehan, G, Mcelroy, A.B, Drewry, D, Lambert, M.H, Jordan, S.R. | | Deposit date: | 1993-11-17 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the catalytic domain of fibroblast collagenase complexed with an inhibitor.
Science, 263, 1994
|
|
6BQP
 
 | | Crystal Structure of the Human CAMKK2B in complex with Crenolanib | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-{5-[(3-Methyloxetan-3-yl)methoxy]-1H-benzimidazol-1-yl}quinolin-8-yl)piperidin-4-amine, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Counago, R.M, de Souza, G.P, dos Reis, C.V, Ramos, P.Z, Drewry, D, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Human CAMKK2B in complex with Crenolanib
To Be Published
|
|
5UYJ
 
 | | Crystal Structure of the Human CAMKK2B | | Descriptor: | 2-cyclopentyl-4-(7-methoxyquinolin-4-yl)benzoic acid, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Counago, R.M, Drewry, D, Arruda, P, Edwards, A.M, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-24 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Human CAMKK2B
To Be Published
|
|
5IKW
 
 | | Crystal Structure of BMP-2-inducible kinase in complex with an Indazole inhibitor | | Descriptor: | BMP-2-inducible protein kinase, N-(6-{3-[(cyclopropylsulfonyl)amino]phenyl}-1H-indazol-3-yl)cyclopropanecarboxamide | | Authors: | Counago, R.M, Sorrell, F.J, Krojer, T, Savitsky, P, Elkins, J.M, Axtman, A, Drewry, D, Wells, C, Zhang, C, Zuercher, W, Willson, T.M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Arruda, P, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-04 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of BMP-2-inducible kinase in complex with a 3-acylaminoindazole inhibitor GSK3236425A
To Be Published
|
|
6BLE
 
 | | Crystal Structure of the Human CAMKK2B in complex with CP673451 | | Descriptor: | 1-{2-[5-(2-methoxyethoxy)-1H-benzimidazol-1-yl]quinolin-8-yl}piperidin-4-amine, Calcium/calmodulin-dependent protein kinase kinase 2, GLYCEROL | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Ramos, P.Z, Drewry, D, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-10 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human CAMKK2B in complex with CP673451
To Be Published
|
|
6BRC
 
 | | Crystal Structure of the Human CAMKK2B in complex with AP26113-analog (ALK-IN-1) | | Descriptor: | 5-chloro-N~2~-{4-[4-(dimethylamino)piperidin-1-yl]-2-methoxyphenyl}-N~4~-[2-(dimethylphosphoryl)phenyl]pyrimidine-2,4-diamine, Calcium/calmodulin-dependent protein kinase kinase 2, SODIUM ION | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Ramos, P.Z, Drewry, D, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-30 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Human CAMKK2B in complex with AP26113-analog (ALK-IN-1)
To Be Published
|
|
6BKU
 
 | | Crystal Structure of the Human CAMKK2B bound to GSK650394 | | Descriptor: | 2-cyclopentyl-4-(5-phenyl-1H-pyrrolo[2,3-b]pyridin-3-yl)benzoic acid, Calcium/calmodulin-dependent protein kinase kinase 2, FORMIC ACID | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Ramos, P.Z, Drewry, D, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-09 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human CAMKK2B bound to GSK650394
To Be Published
|
|
6BQQ
 
 | | Crystal Structure of the Human CAMKK2B in complex with BI2526 | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, ACETATE ION, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Counago, R.M, de Souza, G.P, dos Reis, C.V, Ramos, P.Z, Drewry, D, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Human CAMKK2B in complex with BI2526
To Be Published
|
|
