6NYU
 
 | | The X-ray crystal structure of Staphylococcus aureus Fatty Acid Kinase (Fak) B1 F263T mutant protein to 2.18 Angstrom resolution | | Descriptor: | DegV domain-containing protein, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Cuypers, M.G, Gullett, J.M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | The X-ray crystal structure of STAPHYLOCOCCUS AUREUS Fatty Acid Kinase (Fak) B1 protein F263T mutant to 2.18 Angstrom resolution
To Be Published
|
|
6NR1
 
 | | The X-ray crystal structure of Streptococcus pneumoniae Fatty Acid Kinase (Fak) B2 protein loaded with Vaccenic acid (C18:1 delta11) to 2.1 Angstrom resolution | | Descriptor: | Fatty Acid Kinase (Fak) B2 protein (SPR1019), GLYCEROL, VACCENIC ACID | | Authors: | Cuypers, M.G, Gullett, J.M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2019-01-22 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray crystal structure of Streptococcus pneumoniae Fatty Acid Kinase (Fak) B2 protein loaded with Vaccenic acid (C18:1 delta11) to 2.1 Angstrom resolution
To Be Published
|
|
6W6B
 
 | | The X-ray crystal structure of the C-terminus domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 328-548) protein to 1.40 Angstrom resolution | | Descriptor: | SULFATE ION, SaFakA-Cterminus domain | | Authors: | Cuypers, M.G, Subramanian, C, Rock, C.O, White, S.W. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The X-ray crystal structure of the C-terminus domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 328-548) protein to 1.40 Angstrom resolution
To Be Published
|
|
7RM7
 
 | | The X-ray crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with ADP to 1.025 Angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Fatty Acid Kinase A, GLYCEROL, ... | | Authors: | Cuypers, M.G, Subramanian, C, Rock, C.O, White, S.W. | | Deposit date: | 2021-07-26 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.025 Å) | | Cite: | The X-ray crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with ADP to 1.025 Angstrom resolution
To Be Published
|
|
7RZK
 
 | | Co-soak crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with ADP to 1.9 Angstrom resolution - SAD data | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, Fatty Acid Kinase A, ... | | Authors: | Cuypers, M.G, Subramanian, C, Rock, C.O, White, S.W. | | Deposit date: | 2021-08-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Co-soak crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with ADP to 1.9 Angstrom resolution - SAD data
To Be Published
|
|
6TGT
 
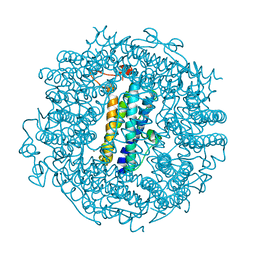 | |
6TB5
 
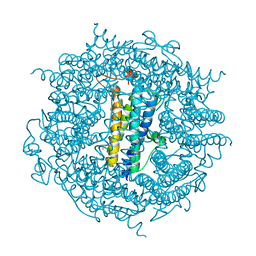 | | The crystal structure of the DPS2 from DEINOCOCCUS RADIODURANS to 1.83A resolution (sequentially soaked in CaCl2 [5mM] for 20 min, then in Ammonium iron(II) sulfate [10mM] for 2h). | | Descriptor: | CALCIUM ION, DNA protection during starvation protein 2, FE (III) ION | | Authors: | Cuypers, M.G, McSweeney, S, Romao, C.V, Mitchell, E.P. | | Deposit date: | 2019-10-31 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of the DPS2 from DEINOCOCCUS RADIODURANS to 1.83A resolution (sequentially soaked in CaCl2 [5mM] for 20 min, then in Ammonium iron(II) sulfate [10mM] for 2h).
To Be Published
|
|
5JLT
 
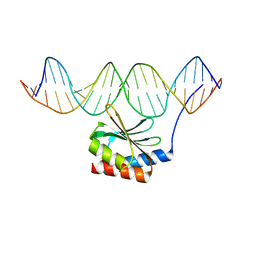 | | The crystal structure of the bacteriophage T4 MotA C-terminal domain in complex with dsDNA reveals a novel protein-DNA recognition motif | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*TP*GP*CP*TP*TP*AP*AP*TP*AP*AP*TP*CP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*GP*AP*TP*TP*AP*TP*TP*AP*AP*GP*CP*AP*AP*AP*GP*CP*TP*TP*C)-3'), Middle transcription regulatory protein motA | | Authors: | Cuypers, M.G, Robertson, R.M, Knipling, L, Hinton, D.M, White, S.W. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | The phage T4 MotA transcription factor contains a novel DNA binding motif that specifically recognizes modified DNA.
Nucleic Acids Res., 46, 2018
|
|
7K77
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ001008025 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-09-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ001008025
To Be Published
|
|
5NW3
 
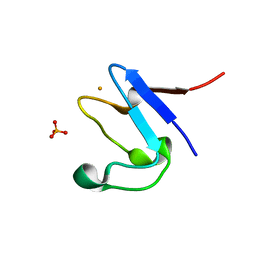 | | The cryofrozen atomic resolution X-ray crystal structure of perdeuterated Pyrococcus furiosus Rubredoxin (100K, 0.59A resolution) | | Descriptor: | FE (III) ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Cuypers, M.G, Mason, S.A, Mossou, E, Haertlein, M, Forsyth, V.T. | | Deposit date: | 2017-05-04 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.59 Å) | | Cite: | The cryofrozen atomic resolution X-ray crystal structure of perdeuterated Pyrococcus furiosus Rubredoxin (100K, 0.59A resolution)
To Be Published
|
|
5OME
 
 | | The cryofrozen atomic resolution X-ray crystal structure of the reduced form (Fe2+) perdeuterated Pyrococcus furiosus Rubredoxin in D2O (100K, 0.75 Angstrom resolution) | | Descriptor: | FE (III) ION, PHOSPHATE ION, Rubredoxin, ... | | Authors: | Cuypers, M.G, Mason, S.A, Mossou, E, Haertlein, M, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2017-07-31 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (0.747 Å) | | Cite: | The cryofrozen atomic resolution X-ray crystal structure of the reduced form (Fe2+) perdeuterated Pyrococcus furiosus Rubredoxin in D2O (100K, 0.75 Angstrom resolution)
To Be Published
|
|
7KNR
 
 | | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988558 | | Descriptor: | 2-(2,6-difluorophenyl)-5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, White, S.W. | | Deposit date: | 2020-11-05 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988558
To Be Published
|
|
7KNY
 
 | | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988528 | | Descriptor: | 2-(2-fluorophenyl)-5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, White, S.W. | | Deposit date: | 2020-11-06 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988528
To Be Published
|
|
7KOP
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease I38T mutant in complex with SJ000988539 | | Descriptor: | 2-(2-fluorophenyl)-5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Seetharaman, J, White, S.W. | | Deposit date: | 2020-11-09 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The crystal structure of the 2009/H1N1/California PA endonuclease I38T mutant in complex with SJ000988539
To Be Published
|
|
7LP8
 
 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476 | | Descriptor: | (phenylmethyl) (2~{S})-2-[5-oxidanyl-6-oxidanylidene-4-(2-pyridin-4-ylethylcarbamoyl)-1~{H}-pyrimidin-2-yl]pyrrolidine-1-carboxylate, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476
To Be Published
|
|
7LP7
 
 | | The crystal structure of the wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476 | | Descriptor: | (phenylmethyl) (2~{S})-2-[5-oxidanyl-6-oxidanylidene-4-(2-pyridin-4-ylethylcarbamoyl)-1~{H}-pyrimidin-2-yl]pyrrolidine-1-carboxylate, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476
To Be Published
|
|
7ML8
 
 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ001023038 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-2-phenyl-1,6-dihydropyrimidine-4-carboxamide, GLUTAMIC ACID, Hexa Vinylpyrrolidone K15, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Yun, M.K, Dubois, R, Rankovic, Z, White, S.W. | | Deposit date: | 2021-04-27 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with St Jude compound AC067-19
To Be Published
|
|
5UTO
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstrom resolution | | Descriptor: | EDD domain protein, DegV family, PALMITIC ACID | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, Broussard, T.C, Miller, D.J, White, S.W, Rock, C.O. | | Deposit date: | 2017-02-15 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstroem resolution
J.Biol.Chem., 2018
|
|
5V85
 
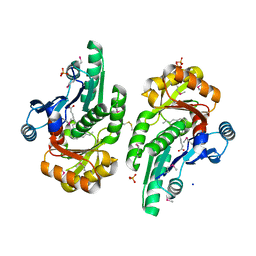 | | The crystal structure of the protein of DegV family COG1307 from Ruminococcus gnavus ATCC 29149 (alternative refinement of PDB 3JR7 with Vaccenic acid) | | Descriptor: | EDD domain protein, DegV family, PHOSPHATE ION, ... | | Authors: | Cuypers, M.G, Ericson, M, subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-03-21 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstroem resolution
J.Biol.Chem., 2018
|
|
5UXY
 
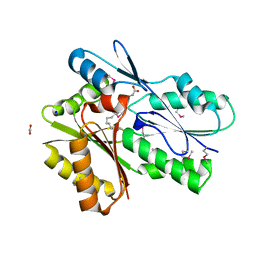 | | The crystal structure of a DegV family protein from Eubacterium eligens loaded with heptadecanoic acid to 1.80 Angstrom resolution (ALTERNATIVE REFINEMENT OF PDB 3FDJ with HEPTADECANOIC acid) | | Descriptor: | ACETIC ACID, DegV family protein, SODIUM ION, ... | | Authors: | Cuypers, M.G, Ericson, M, subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-02-23 | | Release date: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstroem resolution
J.Biol.Chem., 2018
|
|
7MY5
 
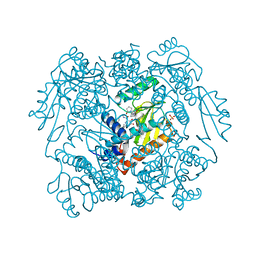 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-05-20 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503
To Be Published
|
|
7MX0
 
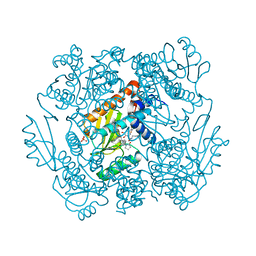 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988558 | | Descriptor: | 2-(2,6-difluorophenyl)-5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-05-17 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988558
To Be Published
|
|
6MH9
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein A121I mutant to 2.02 Angstrom resolution | | Descriptor: | Fatty Acid Kinase (Fak) B1 protein, PALMITIC ACID | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2018-09-17 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
7RKP
 
 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with cyclic compound SJ001034733 | | Descriptor: | (5R)-5-hydroxy-16,21-dioxa-3,8,28-triazatetracyclo[20.3.1.1~2,6~.1~11,15~]octacosa-1(26),2,6(28),11(27),12,14,22,24-octaene-4,7-dione, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
8DTW
 
 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with compound SJ001023036 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-{[2-(trifluoromethyl)phenyl]methyl}-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MAGNESIUM ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2022-07-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
