3KU4
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | SULFATE ION, Uridine phosphorylase | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-26 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
3KUK
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 2'-DEOXYURIDINE, SULFATE ION, Uridine phosphorylase | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
3KVY
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 1,4-anhydro-D-erythro-pent-1-enitol, SULFATE ION, URACIL, ... | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
4MGJ
 
 | | Crystal structure of cytochrome P450 2B4 F429H in complex with 4-CPI | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, Y, Zhang, H, Usharani, D, Bu, W, Im, S, Tarasev, M, Rwere, F, Meagher, J, Sun, C, Stuckey, J, Shaik, S, Waskell, L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural and Functional Characterization of a Cytochrome P450 2B4 F429H Mutant with an Axial Thiolate-Histidine Hydrogen Bond.
Biochemistry, 53, 2014
|
|
3H0M
 
 | | Structure of trna-dependent amidotransferase gatcab from aquifex aeolicus | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, GLUTAMINE, Glutamyl-tRNA(Gln) amidotransferase subunit A, ... | | Authors: | Wu, J, Bu, W, Sheppard, K, Kitabatake, M, Soll, D, Smith, J.L. | | Deposit date: | 2009-04-09 | | Release date: | 2009-07-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into tRNA-Dependent Amidotransferase Evolution and Catalysis from the Structure of the Aquifex aeolicus Enzyme
J.Mol.Biol., 391, 2009
|
|
3H0R
 
 | | Structure of trna-dependent amidotransferase gatcab from aquifex aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASPARAGINE, ... | | Authors: | Wu, J, Bu, W, Sheppard, K, Kitabatake, M, Soll, D, Smith, J.L. | | Deposit date: | 2009-04-10 | | Release date: | 2009-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into tRNA-Dependent Amidotransferase Evolution and Catalysis from the Structure of the Aquifex aeolicus Enzyme
J.Mol.Biol., 391, 2009
|
|
3H0L
 
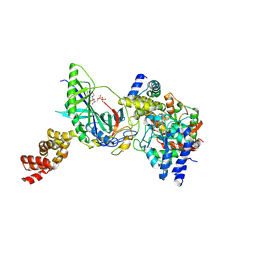 | | Structure of trna-dependent amidotransferase gatcab from aquifex aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ASPARAGINE, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, ... | | Authors: | Wu, J, Bu, W, Sheppard, K, Kitabatake, M, Soll, D, Smith, J.L. | | Deposit date: | 2009-04-09 | | Release date: | 2009-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into tRNA-Dependent Amidotransferase Evolution and Catalysis from the Structure of the Aquifex aeolicus Enzyme
J.Mol.Biol., 391, 2009
|
|
7MWX
 
 | | Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2 with tamarin CD81 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy Chain, ... | | Authors: | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | Deposit date: | 2021-05-17 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
7MWS
 
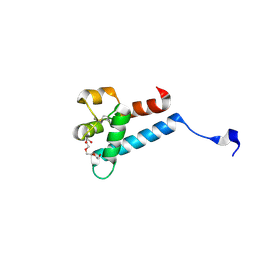 | | Crystal structure of tamarin CD81 large extracellular loop | | Descriptor: | CD81 protein, GLYCEROL, TETRAETHYLENE GLYCOL | | Authors: | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | Deposit date: | 2021-05-17 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
7MWW
 
 | | Structure of hepatitis C virus envelope full-length glycoprotein 2 (eE2) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, ... | | Authors: | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | Deposit date: | 2021-05-17 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
8SM1
 
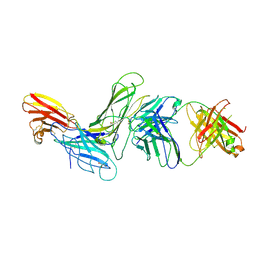 | | CRYSTAL STRUCTURE OF HUMAN ANTIBODY 769A9 IN COMPLEX WITH EPSTEIN-BARR VIRUS MAJOR GLYCOPROTEIN GP350 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769A9 Fab heavy chain, 769A9 Fab light chain, ... | | Authors: | Chen, W.-H, Bu, W, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-01 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural basis for complement receptor engagement and virus neutralization through Epstein-Barr virus gp350.
Immunity, 58, 2025
|
|
8SM0
 
 | | Crystal structure of human complement receptor 2 (CD21) in complex with Epstein-Barr virus major glycoprotein gp350 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement receptor type 2, Envelope glycoprotein gp350, ... | | Authors: | Chen, W.-H, Bu, W, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-01 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for complement receptor engagement and virus neutralization through Epstein-Barr virus gp350.
Immunity, 58, 2025
|
|
5AGS
 
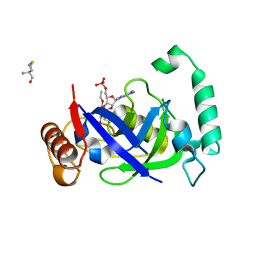 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct 3-(Aminomethyl)-4-bromo-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | Descriptor: | 3-(AMINOMETHYL)-4-BROMO-7-ETHOXYBENZO[C][1,2]OXABOROL-1(3H)-OL-MODIFIED ADENOSINE, LEUCYL-TRNA SYNTHETASE, METHIONINE | | Authors: | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5AGT
 
 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct (S)-3-(Aminomethyl)-4-chloro-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, GLYCEROL, LEUCINE--TRNA LIGASE, ... | | Authors: | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5AGR
 
 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct (S)-3-(Aminomethyl)-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | Descriptor: | 1,2-ETHANEDIOL, 3-AMINOMETHYL-7-(ETHOXY)-3H-BENZO[C][1,2]OXABOROL-1-OL modified adenosine, LEUCINE, ... | | Authors: | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
4L6Q
 
 | | ROCK2 in complex with benzoxaborole | | Descriptor: | 6-[4-(aminomethyl)-2-fluorophenoxy]-2,1-benzoxaborol-1(3H)-ol, Rho-associated protein kinase 2 | | Authors: | Rock, F, Jarnagin, K. | | Deposit date: | 2013-06-12 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Linking phenotype to kinase: identification of a novel benzoxaborole hinge-binding motif for kinase inhibition and development of high-potency rho kinase inhibitors.
J.Pharmacol.Exp.Ther., 347, 2013
|
|
7S0J
 
 | | Crystal structure of Epstein-Barr virus gH/gL targeting antibody 769B10 | | Descriptor: | 769B10 Fab Heavy chain, 769B10 Fab Light chain, GLYCEROL | | Authors: | Chen, W.-H, Kanekiyo, M, Cohen, J.I, Joyce, M.G. | | Deposit date: | 2021-08-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
7S08
 
 | |
7S1B
 
 | | Crystal structure of Epstein-Barr virus glycoproteins gH/gL/gp42-peptide in complex with human neutralizing antibodies 769C2 and 770F7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 Fab heavy chain, 769C2 Fab light chain, ... | | Authors: | Chen, W.-H, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2021-09-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
7S07
 
 | | Crystal structure of Epstein-Barr virus glycoprotein gH/gL/gp42-peptide in complex with human neutralizing antibodies 769B10 and 769C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769B10 Fab heavy chain, 769B10 Fab light chain, ... | | Authors: | Chen, W.-H, Kanekiyo, M, Cohen, J.I, Joyce, M.G. | | Deposit date: | 2021-08-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
6BWW
 
 | |
9DWN
 
 | | hTHIK1 Cryo-EM structure in GDN detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, POTASSIUM ION, Potassium channel subfamily K member 13 | | Authors: | Riel, E.B, Riegelhaupt, P.M. | | Deposit date: | 2024-10-09 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The cryo-EM structure and physical basis for anesthetic inhibition of the THIK1 K2P channel.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8SEF
 
 | | Crystal structure of Cy137C02, a monoclonal antibody isolated from macaques immunized with an Epstein-Barr virus glycoprotein 350 (gp350) nanoparticle vaccine | | Descriptor: | Cy137C02 Fab heavy chain, Cy137C02 Fab light chain, GLYCEROL, ... | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-09 | | Release date: | 2024-04-17 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural basis for complement receptor engagement and virus neutralization through Epstein-Barr virus gp350.
Immunity, 58, 2025
|
|
8SGN
 
 | | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy651H02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-17 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for complement receptor engagement and virus neutralization through Epstein-Barr virus gp350.
Immunity, 58, 2025
|
|
8SGA
 
 | | Crystal structure of 770E11, a monoclonal antibody isolated from a human Epstein-Barr virus seropositive donor | | Descriptor: | 770E11 Fab heavy chain, 770E11 Fab light chain, GLYCEROL, ... | | Authors: | Chen, W.H, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-17 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for complement receptor engagement and virus neutralization through Epstein-Barr virus gp350.
Immunity, 58, 2025
|
|
