2D30
 
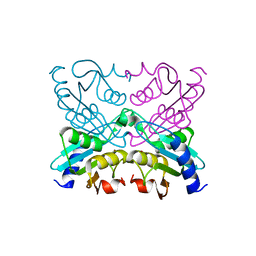 | | Crystal Structure of Cytidine Deaminase Cdd-2 (BA4525) from Bacillus Anthracis at 2.40A Resolution | | Descriptor: | ZINC ION, cytidine deaminase | | Authors: | Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Moroz, O.V, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-09-21 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Cytidine Deaminase Cdd-2 (BA4525) from Bacillus Anthracis at 2.40A Resolution
To be Published
|
|
2C8J
 
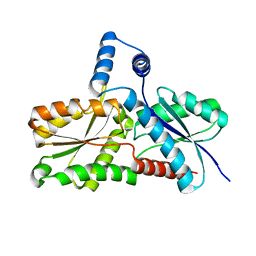 | | CRYSTAL STRUCTURE OF ferrochelatase HemH-1 from Bacillus anthracis, str. Ames | | Descriptor: | FERROCHELATASE 1 | | Authors: | Muller, A, Lebedev, A.A, Moroz, O.V, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-12-05 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Ferrochelatase Hemh-1 from Bacillus Anthracis, Str. Ames
To be Published
|
|
2BTU
 
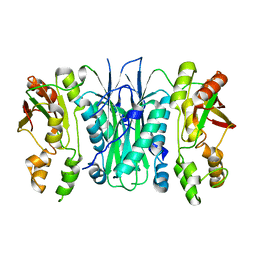 | | Crystal structure of Phosphoribosylformylglycinamidine cyclo-ligase from Bacillus Anthracis at 2.3A resolution. | | Descriptor: | PHOSPHORIBOSYL-AMINOIMIDAZOLE SYNTHETASE | | Authors: | Moroz, O.V, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Lebedev, A.A, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-06-07 | | Release date: | 2006-08-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Phosphoribosylformylglycinamidine Cyclo-Ligase from Bacillus Anthracis at 2.3A Resolution.
To be Published
|
|
2C20
 
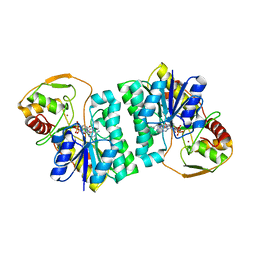 | | CRYSTAL STRUCTURE OF UDP-GLUCOSE 4-EPIMERASE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GLUCOSE 4-EPIMERASE, ZINC ION | | Authors: | Lebedev, A.A, Moroz, O.V, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-09-22 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Udp-Glucose 4-Epimerase from Bacillus Anthracis at 2.7A Resolution
To be Published
|
|
2C40
 
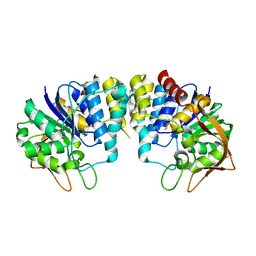 | | CRYSTAL STRUCTURE OF INOSINE-URIDINE PREFERRING NUCLEOSIDE HYDROLASE FROM BACILLUS ANTHRACIS AT 2.2A RESOLUTION | | Descriptor: | CALCIUM ION, INOSINE-URIDINE PREFERRING NUCLEOSIDE HYDROLASE FAMILY PROTEIN, alpha-D-ribofuranose | | Authors: | Moroz, O.V, Blagova, E.V, Fogg, M.J, Levdikov, V.M, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-10-13 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Inosine-Uridine Preferring Nucleoside Hydrolase from Bacillus Anthracis at 2.2A Resolution
To be Published
|
|
5AEO
 
 | | Virulence-associated protein VapG from the intracellular pathogen Rhodococcus equi | | Descriptor: | POTASSIUM ION, R. EQUI VAPG PROTEIN | | Authors: | Okoko, T, Blagova, E.V, Whittingham, J.L, Dover, L.G, Wilkinson, A.J. | | Deposit date: | 2015-01-07 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Characterisation of the Virulence-Associated Protein Vapg from the Horse Pathogen Rhodococcus Equi.
Vet.Microbiol., 179, 2015
|
|
1XE3
 
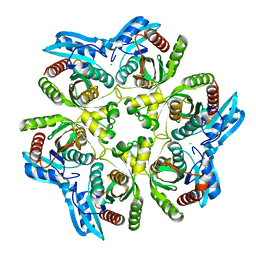 | | Crystal Structure of purine nucleoside phosphorylase DeoD from Bacillus anthracis | | Descriptor: | CHLORIDE ION, purine nucleoside phosphorylase | | Authors: | Grenha, R, Levdikov, V.M, Fogg, M, Blagova, E.V, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-09-09 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure of purine nucleoside phosphorylase (DeoD) from Bacillus anthracis.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1YPF
 
 | | Crystal Structure of GuaC (BA5705) from Bacillus anthracis at 1.8 A Resolution | | Descriptor: | GMP reductase | | Authors: | Grenha, R, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of GuaC (BA5705) from Bacillus anthracis at 1.8 A Resolution.
To be Published
|
|
1XRE
 
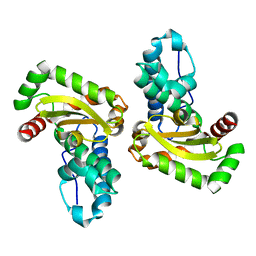 | | Crystal Structure of SodA-2 (BA5696) from Bacillus anthracis at 1.8A Resolution. | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Boucher, I.W, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2004-10-14 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of two superoxide dismutases from Bacillus anthracis reveal a novel active centre.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1XP3
 
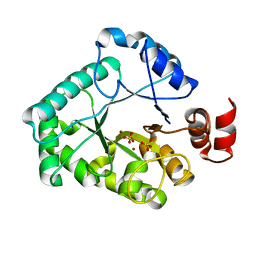 | | Crystal Structure of Endonuclease IV (BA4508) from Bacillus anthracis at 2.57A Resolution. | | Descriptor: | SULFATE ION, ZINC ION, endonuclease IV | | Authors: | Fogg, M.J, Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure of Endonuclease IV (BA4508) from Bacillus anthracis at 2.57A Resolution.
To be Published
|
|
1XUQ
 
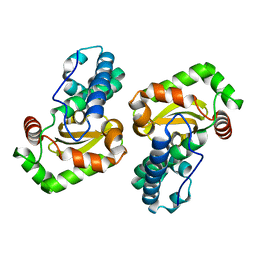 | | Crystal Structure of SodA-1 (BA4499) from Bacillus anthracis at 1.8A Resolution. | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Boucher, I.W, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2004-10-26 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of two superoxide dismutases from Bacillus anthracis reveal a novel active centre.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2A1Y
 
 | | Crystal Structure of GuaC-GMP complex from Bacillus anthracis at 2.26 A Resolution. | | Descriptor: | GMP reductase, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Grenha, R, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of GuaC-GMP complex from Bacillus anthracis at 2.26 A resolution.
To be Published
|
|
5M1Q
 
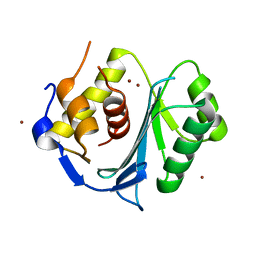 | | Crystal structure of the large terminase nuclease from thermophilic phage G20c with bound Zinc | | Descriptor: | Phage terminase large subunit, ZINC ION | | Authors: | Xu, R.G, Jenkins, H.T, Chechik, M, Blagova, E.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2016-10-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Viral genome packaging terminase cleaves DNA using the canonical RuvC-like two-metal catalysis mechanism.
Nucleic Acids Res., 45, 2017
|
|
5M1K
 
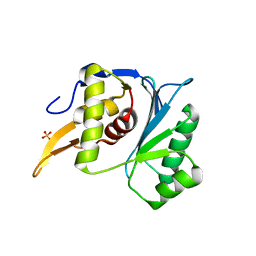 | | Crystal structure of the large terminase nuclease from thermophilic phage G20c with bound Magnesium | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Phage terminase large subunit, ... | | Authors: | Xu, R.G, Jenkins, H.T, Chechik, M, Blagova, E.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2016-10-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Viral genome packaging terminase cleaves DNA using the canonical RuvC-like two-metal catalysis mechanism.
Nucleic Acids Res., 45, 2017
|
|
5M1P
 
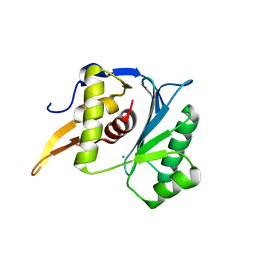 | | Crystal structure of the large terminase nuclease from thermophilic phage G20c with bound Calcium | | Descriptor: | CALCIUM ION, Terminase large subunit | | Authors: | Xu, R.G, Jenkins, H.T, Chechik, M, Blagova, E.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2016-10-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Viral genome packaging terminase cleaves DNA using the canonical RuvC-like two-metal catalysis mechanism.
Nucleic Acids Res., 45, 2017
|
|
5MBU
 
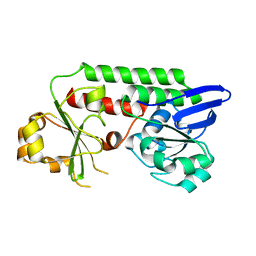 | | CeuE (H227A, Y288F variant) a periplasmic protein from Campylobacter jejuni | | Descriptor: | Enterochelin uptake periplasmic binding protein | | Authors: | Wilde, E.J, Blagova, E.V, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
5MBQ
 
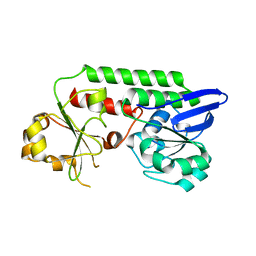 | | CeuE (H227A variant) a periplasmic protein from Campylobacter jejuni | | Descriptor: | Enterochelin uptake periplasmic binding protein | | Authors: | Wilde, E.J, Blagova, E.V, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
5MBT
 
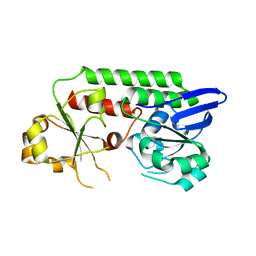 | | CeuE (H227L, Y288F variant) a periplasmic protein from Campylobacter jejuni | | Descriptor: | Enterochelin uptake periplasmic binding protein | | Authors: | Wilde, E.J, Blagova, E.V, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
5M1N
 
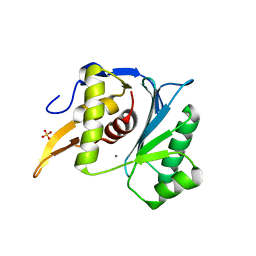 | | Crystal structure of the large terminase nuclease from thermophilic phage G20c with bound Manganese | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, Phage terminase large subunit, ... | | Authors: | Xu, R.G, Jenkins, H.T, Chechik, M, Blagova, E.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2016-10-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Viral genome packaging terminase cleaves DNA using the canonical RuvC-like two-metal catalysis mechanism.
Nucleic Acids Res., 45, 2017
|
|
5MQH
 
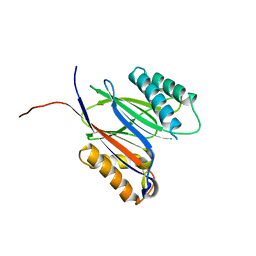 | |
3O9P
 
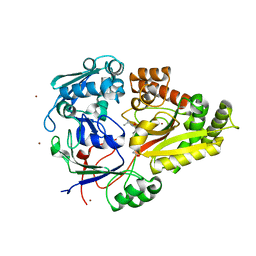 | | The structure of the Escherichia coli murein tripeptide binding protein MppA | | Descriptor: | L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, Periplasmic murein peptide-binding protein, ZINC ION | | Authors: | Maqbool, A, Levdikov, V.M, Blagova, E.V, Wilkinson, A.J, Thomas, G.H. | | Deposit date: | 2010-08-04 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Compensating Stereochemical Changes Allow Murein Tripeptide to Be Accommodated in a Conventional Peptide-binding Protein.
J.Biol.Chem., 286, 2011
|
|
5TCY
 
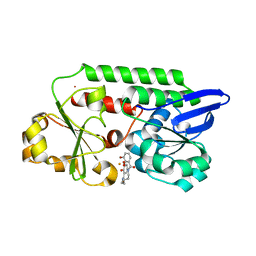 | | A complex of the synthetic siderophore analogue Fe(III)-5-LICAM with CeuE (H227L variant), a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin uptake periplasmic binding protein, FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide) | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
5LOJ
 
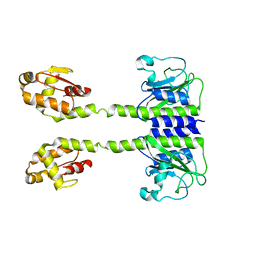 | |
5LOE
 
 | | Structure of full length Cody from Bacillus subtilis in complex with Ile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, ISOLEUCINE | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2016-08-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Branched-chain Amino Acid and GTP-sensing Global Regulator, CodY, from Bacillus subtilis.
J. Biol. Chem., 292, 2017
|
|
5LNH
 
 | | Structure of full length Unliganded CodY from Bacillus subtilis | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, SULFATE ION | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2016-08-04 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Branched-chain Amino Acid and GTP-sensing Global Regulator, CodY, from Bacillus subtilis.
J. Biol. Chem., 292, 2017
|
|
