2NWO
 
 | |
2NXS
 
 | |
2NWY
 
 | |
2NWZ
 
 | |
2NXR
 
 | |
2NWP
 
 | |
2NXT
 
 | |
1L7B
 
 | | Solution NMR Structure of BRCT Domain of T. Thermophilus: Northeast Structural Genomics Consortium Target WR64TT | | Descriptor: | DNA LIGASE | | Authors: | Sahota, G, Dixon, B.L, Huang, Y.P, Aramini, J, Monleon, D, Bhattacharya, D, Swapna, G.V.T, Yin, C, Xiao, R, Anderson, S, Tejero, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-03-14 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Brct Domain from Thermus Thermophilus DNA Ligase
To be Published
|
|
6W5D
 
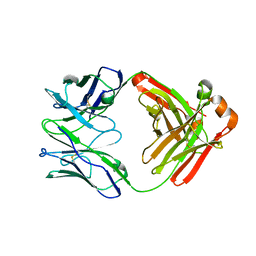 | | Crystal Structure of Fab RSB1 | | Descriptor: | RSB1 Fab Heavy Chain, RSB1 Fab Light Chain | | Authors: | Harshbarger, W, Chandramouli, S, Malito, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Convergent structural features of respiratory syncytial virus neutralizing antibodies and plasticity of the site V epitope on prefusion F.
Plos Pathog., 16, 2020
|
|
6W52
 
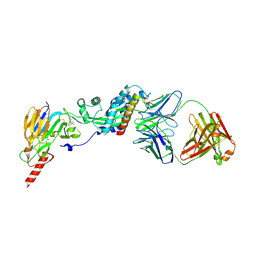 | | Prefusion RSV F bound by neutralizing antibody RSB1 | | Descriptor: | Fusion glycoprotein F0, Fusion glycoprotein F1 fused with Fibritin trimerization domain, RSB1 Fab Heavy Chain, ... | | Authors: | Harshbarger, W, Chandramouli, S, Malito, M. | | Deposit date: | 2020-03-12 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Convergent structural features of respiratory syncytial virus neutralizing antibodies and plasticity of the site V epitope on prefusion F.
Plos Pathog., 16, 2020
|
|
3EOV
 
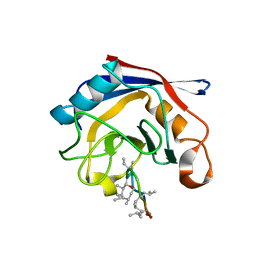 | | Crystal structure of cyclophilin from Leishmania donovani ligated with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Venugopal, V, Dasgupta, D, Datta, A.K, Banerjee, R. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Cyclophilin from Leishmania Donovani Bound to Cyclosporin at 2.6 A Resolution: Correlation between Structure and Thermodynamic Data.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5YKL
 
 | |
5YKQ
 
 | |
5YKK
 
 | |
8E2B
 
 | | N-terminal domain of S. aureus GpsB | | Descriptor: | Cell cycle protein GpsB, GLYCEROL | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2022-08-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Staphylococcus aureus FtsZ and PBP4 bind to the conformationally dynamic N-terminal domain of GpsB.
Elife, 13, 2024
|
|
8E2C
 
 | |
5XES
 
 | | TK9 NMR structure in SDS micelle | | Descriptor: | THR-VAL-TYR-VAL-TYR-SER-ARG-VAL-LYS | | Authors: | Ghosh, A, Bhunia, A. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights of a self-assembling 9-residue peptide from the C-terminal tail of the SARS corona virus E-protein in DPC and SDS micelles: A combined high and low resolution spectroscopic study.
Biochim Biophys Acta Biomembr, 1860, 2018
|
|
5XER
 
 | | TK9 NMR structure in DPC micelle | | Descriptor: | THR-VAL-TYR-VAL-TYR-SER-ARG-VAL-LYS | | Authors: | Ghosh, A, Bhunia, A. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights of a self-assembling 9-residue peptide from the C-terminal tail of the SARS corona virus E-protein in DPC and SDS micelles: A combined high and low resolution spectroscopic study.
Biochim Biophys Acta Biomembr, 1860, 2018
|
|
6JPV
 
 | | Structural analysis of AIMP2-DX2 and HSP70 interaction | | Descriptor: | Heat shock 70 kDa protein 1A,Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 | | Authors: | Cho, H.Y, Son, S.Y, Jeon, Y.H. | | Deposit date: | 2019-03-28 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15000653 Å) | | Cite: | Targeting the interaction of AIMP2-DX2 with HSP70 suppresses cancer development.
Nat.Chem.Biol., 16, 2020
|
|
6K39
 
 | | Structural analysis of AIMP2-DX2 and HSP70 interaction | | Descriptor: | Heat shock 70 kDa protein 1A,Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 | | Authors: | Cho, H.Y, Son, S.Y, Jeon, Y.H. | | Deposit date: | 2019-05-16 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3981427 Å) | | Cite: | Targeting the interaction of AIMP2-DX2 with HSP70 suppresses cancer development.
Nat.Chem.Biol., 16, 2020
|
|
2WF7
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphonate and Aluminium tetrafluoride | | Descriptor: | 6,7-dideoxy-7-phosphono-beta-D-gluco-heptopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Alpha-Fluorophosphonates Reveal How a Phosphomutase Conserves Transition State Conformation Over Hexose Recognition in its Two-Step Reaction.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4C4S
 
 | |
4C4R
 
 | |
4C4T
 
 | |
5OLW
 
 | |
