1OO2
 
 | | Crystal structure of transthyretin from Sparus aurata | | Descriptor: | CADMIUM ION, transthyretin | | Authors: | Pasquato, N, Ramazzina, I, Folli, C, Battistutta, R, Berni, R, Zanotti, G. | | Deposit date: | 2003-03-03 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Distinctive binding and structural properties of piscine transthyretin.
Febs Lett., 555, 2003
|
|
1FEM
 
 | | CRYSTALLOGRAPHIC STUDIES ON COMPLEXES BETWEEN RETINOIDS AND PLASMA RETINOL-BINDING PROTEIN | | Descriptor: | RETINOIC ACID, RETINOL BINDING PROTEIN | | Authors: | Zanotti, G, Marcello, M, Malpeli, G, Sartori, G, Berni, R. | | Deposit date: | 1994-08-29 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic studies on complexes between retinoids and plasma retinol-binding protein.
J.Biol.Chem., 269, 1994
|
|
1FEN
 
 | | CRYSTALLOGRAPHIC STUDIES ON COMPLEXES BETWEEN RETINOIDS AND PLASMA RETINOL-BINDING PROTEIN | | Descriptor: | ALL-TRANS AXEROPHTHENE, RETINOL BINDING PROTEIN | | Authors: | Zanotti, G, Marcello, M, Malpeli, G, Sartori, G, Berni, R. | | Deposit date: | 1994-08-29 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic studies on complexes between retinoids and plasma retinol-binding protein.
J.Biol.Chem., 269, 1994
|
|
4ALO
 
 | | STRUCTURE AND PROPERTIES OF H1 CRUSTACYANIN FROM LOBSTER HOMARUS AMERICANUS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, H1 APOCRUSTACYANIN, SODIUM ION, ... | | Authors: | Ferrari, M, Folli, C, Pincolini, E, Mcclintock, T.S, Roessle, M, Berni, R, Cianci, M. | | Deposit date: | 2012-03-05 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Characterization of Recombinant Crustacyanin Subunits from the Lobster Homarus Americanus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5M98
 
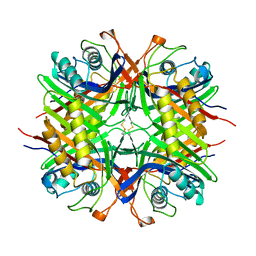 | |
2B5S
 
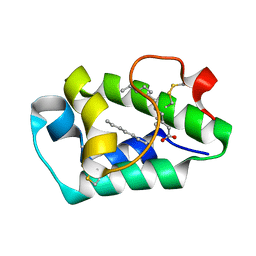 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, LAURIC ACID, Non-specific lipid transfer protein, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, R.J, Zanotti, G. | | Deposit date: | 2005-09-29 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
5LL1
 
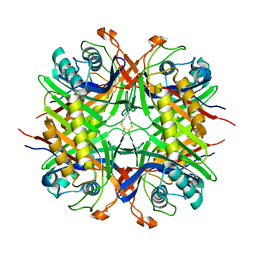 | |
1BOI
 
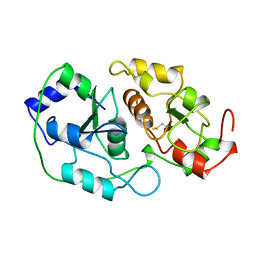 | | N-TERMINALLY TRUNCATED RHODANESE | | Descriptor: | RHODANESE | | Authors: | Gliubich, F, Berni, R, Cianci, M, Trevino, R.J, Horowitz, P.M, Zanotti, G. | | Deposit date: | 1998-08-04 | | Release date: | 1999-04-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | NH2-terminal sequence truncation decreases the stability of bovine rhodanese, minimally perturbs its crystal structure, and enhances interaction with GroEL under native conditions.
J.Biol.Chem., 274, 1999
|
|
1BOH
 
 | | SULFUR-SUBSTITUTED RHODANESE (ORTHORHOMBIC FORM) | | Descriptor: | RHODANESE | | Authors: | Gliubich, F, Berni, R, Cianci, M, Trevino, R.J, Horowitz, P.M, Zanotti, G. | | Deposit date: | 1998-08-04 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NH2-terminal sequence truncation decreases the stability of bovine rhodanese, minimally perturbs its crystal structure, and enhances interaction with GroEL under native conditions.
J.Biol.Chem., 274, 1999
|
|
1AQB
 
 | | RETINOL-BINDING PROTEIN (RBP) FROM PIG PLASMA | | Descriptor: | CADMIUM ION, RETINOL, RETINOL-BINDING PROTEIN | | Authors: | Zanotti, G, Panzalorto, M, Marcato, A, Malpeli, G, Folli, C, Berni, R. | | Deposit date: | 1997-07-28 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of pig plasma retinol-binding protein at 1.65 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2ALG
 
 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, HEXAETHYLENE GLYCOL, LAURIC ACID, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, J, Zanotti, G. | | Deposit date: | 2005-08-05 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
3ISL
 
 | | Crystal structure of ureidoglycine-glyoxylate aminotransferase (pucG) from Bacillus subtilis | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Purine catabolism protein pucG | | Authors: | Costa, R, Cendron, L, Ramazzina, I, Berni, R, Peracchi, A, Percudani, R, Zanotti, G. | | Deposit date: | 2009-08-26 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Amino acids from purines in GUT bacteria
To be Published
|
|
6SUG
 
 | | Crystal structure of transthyretin in complex with 3-deoxytolcapone, a tolcapone analogue | | Descriptor: | 3-deoxytolcapone, Transthyretin | | Authors: | Loconte, V, Cianci, M, Menozzi, I, Sbravati, D, Sansone, F, Casnati, A, Berni, R. | | Deposit date: | 2019-09-14 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Interactions of tolcapone analogues as stabilizers of the amyloidogenic protein transthyretin.
Bioorg.Chem., 103, 2020
|
|
1QQF
 
 | | N-TERMINALLY TRUNCATED C3D,G FRAGMENT OF THE COMPLEMENT SYSTEM | | Descriptor: | PROTEIN (COMPLEMENT C3DG) | | Authors: | Zanotti, G, Bassetto, A, Battistutta, R, Stoppini, M, Berni, R. | | Deposit date: | 1999-06-04 | | Release date: | 2000-07-31 | | Last modified: | 2015-01-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure at 1.44 A resolution of an N-terminally truncated form of the rat serum complement C3d fragment.
Biochim.Biophys.Acta, 1478, 2000
|
|
6SUH
 
 | | Crystal structure of human transthyretin in complex with 3-O-methyltolcapone, a tolcapone analogue | | Descriptor: | 3-O-methyltolcapone, Transthyretin | | Authors: | Loconte, V, Cianci, M, Menozzi, I, Sbravati, D, Sansone, F, Casnati, A, Berni, R. | | Deposit date: | 2019-09-14 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Interactions of tolcapone analogues as stabilizers of the amyloidogenic protein transthyretin.
Bioorg.Chem., 103, 2020
|
|
1QSJ
 
 | | N-TERMINALLY TRUNCATED C3DG FRAGMENT | | Descriptor: | COMPLEMENT C3 PRECURSOR | | Authors: | Zanotti, G, Bassetto, A, Battistutta, R, Stoppini, M, Folli, C, Berni, R. | | Deposit date: | 1999-06-22 | | Release date: | 2000-07-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure at 1.44 A resolution of an N-terminally truncated form of the rat serum complement C3d fragment.
Biochim.Biophys.Acta, 1478, 2000
|
|
5LJE
 
 | | Crystal structure of holo human CRBP1/K40L,Q108L mutant | | Descriptor: | RETINOL, Retinol-binding protein 1, SODIUM ION | | Authors: | Zanotti, G, Vallese, F, Berni, R, Menozzi, I. | | Deposit date: | 2016-07-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and molecular determinants affecting the interaction of retinol with human CRBP1.
J. Struct. Biol., 197, 2017
|
|
1KT5
 
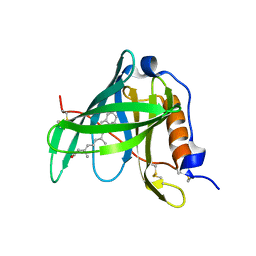 | | Crystal structure of bovine holo-RBP at pH 4.0 | | Descriptor: | Plasma retinol-binding protein, RETINOL | | Authors: | Calderone, V, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | High-resolution Structures of Retinol-binding Protein in
Complex with Retinol: pH-induced Protein Structural
Changes in the Crystal State
J.Mol.Biol., 329, 2003
|
|
2O70
 
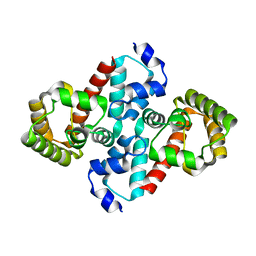 | | Structure of OHCU decarboxylase from zebrafish | | Descriptor: | OHCU decarboxylase | | Authors: | Cendron, L, Berni, R, Folli, C, Ramazzina, I, Percudani, R, Zanotti, G. | | Deposit date: | 2006-12-09 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
J.Biol.Chem., 282, 2007
|
|
2O73
 
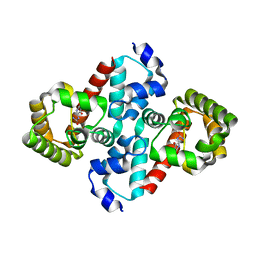 | | Structure of OHCU decarboxylase in complex with allantoin | | Descriptor: | 1-(2,5-DIOXO-2,5-DIHYDRO-1H-IMIDAZOL-4-YL)UREA, OHCU decarboxylase | | Authors: | Cendron, L, Berni, R, Folli, C, Ramazzina, I, Percudani, R, Zanotti, G. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
J.Biol.Chem., 282, 2007
|
|
2O74
 
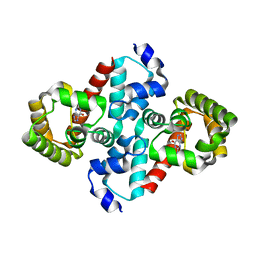 | | Structure of OHCU decarboxylase in complex with guanine | | Descriptor: | GUANINE, OHCU decarboxylase | | Authors: | Cendron, L, Berni, R, Folli, C, Ramazzina, I, Percudani, R, Zanotti, G. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
J.Biol.Chem., 282, 2007
|
|
6R66
 
 | | Crystal structure of transthyretin in complex with CHF5075, a flurbiprofen analogue | | Descriptor: | 1-[4-[3,5-bis(chloranyl)phenyl]-3-fluoranyl-phenyl]cyclopropane-1-carboxylic acid, Transthyretin | | Authors: | Loconte, V, Menozzi, I, Ferrari, A, Berni, R, Zanotti, G. | | Deposit date: | 2019-03-26 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-activity relationships of flurbiprofen analogues as stabilizers of the amyloidogenic protein transthyretin.
J.Struct.Biol., 208, 2019
|
|
6R67
 
 | | Crystal structure of transthyretin in complex with CHF5075, a flurbiprofen analogue | | Descriptor: | (2~{R})-2-[4-(3,5-dimethylphenyl)-3-fluoranyl-phenyl]propanoic acid, Transthyretin | | Authors: | Loconte, V, Menozzi, I, Ferrari, A, Berni, R, Zanotti, G. | | Deposit date: | 2019-03-26 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-activity relationships of flurbiprofen analogues as stabilizers of the amyloidogenic protein transthyretin.
J.Struct.Biol., 208, 2019
|
|
6R68
 
 | | Crystal structure of transthyretin in complex with CHF4795, a flurbiprofen analogue | | Descriptor: | (2~{R})-2-[4-[3,5-bis(chloranyl)phenyl]-3-fluoranyl-phenyl]propanoic acid, Transthyretin | | Authors: | Loconte, V, Menozzi, I, Ferrari, A, Berni, R, Zanotti, G. | | Deposit date: | 2019-03-26 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-activity relationships of flurbiprofen analogues as stabilizers of the amyloidogenic protein transthyretin.
J.Struct.Biol., 208, 2019
|
|
6R6I
 
 | | Crystal structure of transthyretin mutant A25T in complex with CHF5074, a flurbiprofen analogue | | Descriptor: | 1-(3',4'-dichloro-2-fluorobiphenyl-4-yl)cyclopropanecarboxylic acid, PHOSPHATE ION, Transthyretin | | Authors: | Loconte, V, Menozzi, I, Ferrari, A, Berni, R, Zanotti, G. | | Deposit date: | 2019-03-27 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.467 Å) | | Cite: | Structure-activity relationships of flurbiprofen analogues as stabilizers of the amyloidogenic protein transthyretin.
J.Struct.Biol., 208, 2019
|
|
