7UAX
 
 | | The crystal structure of the K36A/K38A double mutant of E. coli YGGS in complex with PLP | | Descriptor: | PHOSPHATE ION, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7U9H
 
 | | Crystal Structure of Escherichia coli apo Pyridoxal 5'-phosphate homeostasis protein (YGGS) | | Descriptor: | Pyridoxal phosphate homeostasis protein, SULFATE ION | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-10 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UAU
 
 | | The crystal structure of the K137A mutant of E. coli YGGS in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein, SULFATE ION | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UBP
 
 | | The crystal structure of the K36A/K137A double mutant of E. coli YGGS in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein, SULFATE ION | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-15 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7U9C
 
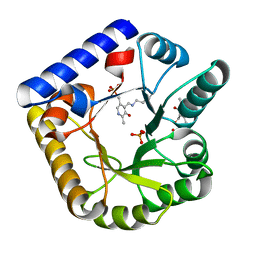 | | Crystal Structure of the wild type Escherichia coli Pyridoxal 5'-phosphate homeostasis protein (YGGS) | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Safo, M.K, Musayev, F.N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UAT
 
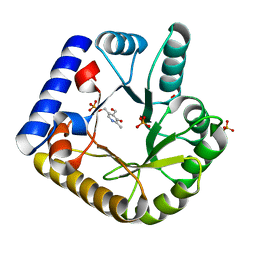 | | The crystal structure of the K36A mutant of E. coli YGGS in complex with PLP | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UB8
 
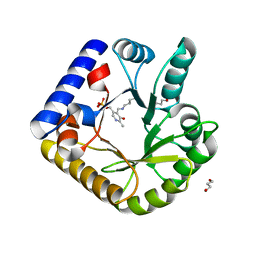 | | The crystal structure of the K38A/K137A/K233A/K234A quadruple mutant of E. coli YGGS in complex with PLP | | Descriptor: | 1,4-BUTANEDIOL, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UBQ
 
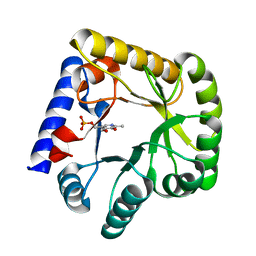 | | The crystal structure of the wild-type of E. coli YGGS in complex with PNP | | Descriptor: | PYRIDOXINE-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-15 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UB4
 
 | | The crystal structure of the K36A/K38A/K233A/K234A quadruple mutant of E. coli YGGS in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
3N4S
 
 | | Structure of Csm1 C-terminal domain, P21212 form | | Descriptor: | Monopolin complex subunit CSM1, PENTAETHYLENE GLYCOL | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-22 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N7N
 
 | | Structure of Csm1/Lrs4 complex | | Descriptor: | Monopolin complex subunit CSM1, Monopolin complex subunit LRS4 | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N4R
 
 | | Structure of Csm1 C-terminal domain, R3 form | | Descriptor: | MALONATE ION, Monopolin complex subunit CSM1, PENTAETHYLENE GLYCOL | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-22 | | Release date: | 2010-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N4X
 
 | | Structure of Csm1 full-length | | Descriptor: | Monopolin complex subunit CSM1 | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-23 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
8BVG
 
 | |
2P1Y
 
 | |
1TRH
 
 | |
2P24
 
 | | I-Au/MBP125-135 | | Descriptor: | H-2 class II histocompatibility antigen, A-U alpha chain, A-U beta chain | | Authors: | McBeth, C, Strong, R.K. | | Deposit date: | 2007-03-06 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A new twist in TCR diversity revealed by a forbidden alphabeta TCR.
J.Mol.Biol., 375, 2008
|
|
8QYW
 
 | |
8QYT
 
 | | Human Pyridoxine-5'-phosphate oxidase in complex with PLP | | Descriptor: | BETA-MERCAPTOETHANOL, FLAVIN MONONUCLEOTIDE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Antonelli, L, Ilari, A, Fiorillo, A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Identification of the pyridoxal 5'-phosphate allosteric site in human pyridox(am)ine 5'-phosphate oxidase.
Protein Sci., 33, 2024
|
|
6FL5
 
 | | Structure of human SHMT1-H135N-R137A-E168N mutant at 3.6 Ang. resolution | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, ... | | Authors: | Giardina, G, Cutruzzola, F, Lucchi, R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The catalytic activity of serine hydroxymethyltransferase is essential for de novo nuclear dTMP synthesis in lung cancer cells.
FEBS J., 285, 2018
|
|
8A11
 
 | | Cryo-EM structure of the Human SHMT1-RNA complex | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, cytosolic | | Authors: | Spizzichino, S, Marabelli, C, Bharadwaj, A, Jakobi, A.J, Chaves-Sanjuan, A, Giardina, G, Bolognesi, M, Cutruzzola, F. | | Deposit date: | 2022-05-30 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structure-based mechanism of riboregulation of the metabolic enzyme SHMT1.
Mol.Cell, 2024
|
|
8R7H
 
 | | Cryo-EM structure of Human SHMT1 | | Descriptor: | Serine hydroxymethyltransferase, cytosolic | | Authors: | Spizzichino, S, Marabelli, C, Bharadwaj, A, Jakobi, A.J, Chaves-Sanjuan, A, Giardina, G, Bolognesi, M, Cutruzzola, F. | | Deposit date: | 2023-11-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure-based mechanism of riboregulation of the metabolic enzyme SHMT1.
Mol.Cell, 84, 2024
|
|
7PQ9
 
 | | Crystal structure of Bacillus clausii pdxR at 2.8 Angstroms resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vivoli Vega, M, Isupov, M.N, Harmer, N. | | Deposit date: | 2021-09-16 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
8PUM
 
 | |
8PUS
 
 | |
