7DR2
 
 | | Structure of GraFix PSI tetramer from Cyanophora paradoxa | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2020-12-25 | | Release date: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into an evolutionary turning-point of photosystem I from prokaryotes to eukaryotes
Biorxiv, 2022
|
|
7DR1
 
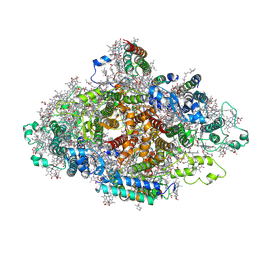 | | Structure of Wild-type PSI monomer2 from Cyanophora paradoxa | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2020-12-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into an evolutionary turning-point of photosystem I from prokaryotes to eukaryotes
Biorxiv, 2022
|
|
7DR0
 
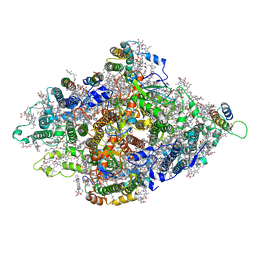 | | Structure of Wild-type PSI monomer1 from Cyanophora paradoxa | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2020-12-25 | | Release date: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into an evolutionary turning-point of photosystem I from prokaryotes to eukaryotes
Biorxiv, 2022
|
|
7F2P
 
 | | The head structure of Helicobacter pylori bacteriophage KHP40 | | Descriptor: | Cement protein gp16, KHP40 MCP | | Authors: | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2021-06-13 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
6KNB
 
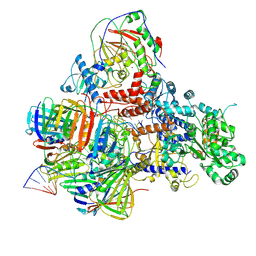 | | PolD-PCNA-DNA (form A) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
6LXV
 
 | | Cryo-EM structure of phosphoketolase from Bifidobacterium longum | | Descriptor: | CALCIUM ION, Phosphoketolase, THIAMINE DIPHOSPHATE | | Authors: | Nakata, K, Miyazaki, N, Yamaguchi, H, Hirose, M, Miyano, H, Mizukoshi, T, Kashiwagi, T, Iwasaki, K. | | Deposit date: | 2020-02-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | High-resolution structure of phosphoketolase from Bifidobacterium longum determined by cryo-EM single-particle analysis.
J.Struct.Biol., 214, 2022
|
|
1UF2
 
 | | The Atomic Structure of Rice dwarf Virus (RDV) | | Descriptor: | Core protein P3, Outer capsid protein P8, Structural protein P7 | | Authors: | Nakagawa, A, Miyazaki, N, Taka, J, Naitow, H, Ogawa, A, Fujimoto, Z, Mizuno, H, Higashi, T, Watanabe, Y, Omura, T, Cheng, R.H, Tsukihara, T. | | Deposit date: | 2003-05-23 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins.
Structure, 11, 2003
|
|
7CEC
 
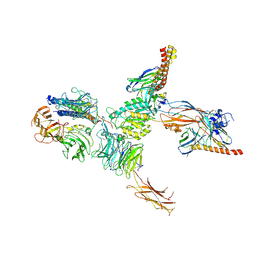 | | Structure of alpha6beta1 integrin in complex with laminin-511 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Arimori, T, Miyazaki, N, Takagi, J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural mechanism of laminin recognition by integrin.
Nat Commun, 12, 2021
|
|
7X30
 
 | | Capsid structure of Staphylococcus jumbo bacteriophage S6 | | Descriptor: | Hoc-like protein ORF90, Major structural protein ORF12 | | Authors: | Koibuchi, W, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2022-02-27 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Capsid structure of Staphylococcus jumbo bacteriophage S6
To Be Published
|
|
7DOU
 
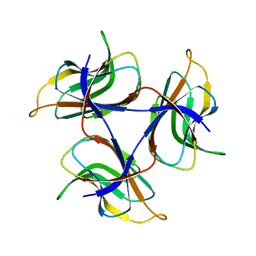 | | Trimeric cement protein structure of Helicobacter pylori bacteriophage KHP40 | | Descriptor: | Cement protein gp16 | | Authors: | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
7DN2
 
 | | Acidic stable capsid structure of Helicobacter pylori bacteriophage KHP30 | | Descriptor: | Cement protein gp15, Major structural protein ORF14 | | Authors: | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
7WMP
 
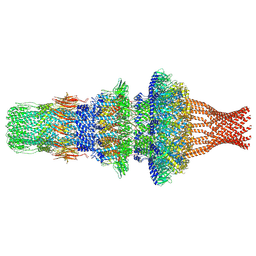 | | Tail structure of Helicobacter pylori bacteriophage KHP30 | | Descriptor: | Adaptor protein gp12, Nozzle protein gp25, Portal protein | | Authors: | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2022-01-15 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori bacteriophage KHP30
To Be Published
|
|
7D1T
 
 | | Cryo-EM Structure of PSII at 1.95 angstrom resolution | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kato, K, Miyazaki, N, Hamaguchi, T, Nakajima, Y, Akita, F, Yonekura, K, Shen, J.R. | | Deposit date: | 2020-09-15 | | Release date: | 2021-03-31 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (1.95 Å) | | Cite: | High-resolution cryo-EM structure of photosystem II reveals damage from high-dose electron beams.
Commun Biol, 4, 2021
|
|
7D1U
 
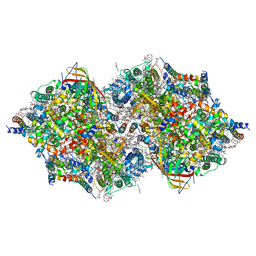 | | Cryo-EM Structure of PSII at 2.08 angstrom resolution | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kato, K, Miyazaki, N, Hamaguchi, T, Nakajima, Y, Akita, F, Yonekura, K, Shen, J.R. | | Deposit date: | 2020-09-15 | | Release date: | 2021-03-31 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | High-resolution cryo-EM structure of photosystem II reveals damage from high-dose electron beams.
Commun Biol, 4, 2021
|
|
3VKF
 
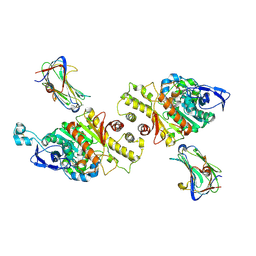 | | Crystal Structure of Neurexin 1beta/Neuroligin 1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurexin-1-beta, ... | | Authors: | Tanaka, H, Miyazaki, N, Nogi, T, Iwasaki, K, Takagi, J. | | Deposit date: | 2011-11-15 | | Release date: | 2012-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Higher-order architecture of cell adhesion mediated by polymorphic synaptic adhesion molecules neurexin and neuroligin.
Cell Rep, 2, 2012
|
|
5GJE
 
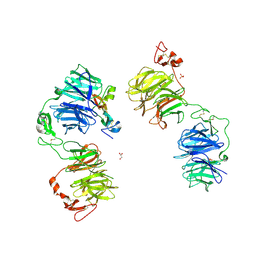 | | Three-dimensional reconstruction of human LRP6 ectodomain complexed with Dkk1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Matoba, K, Mihara, E, Tamura-Kawakami, K, Hirai, H, Thompson, S, Iwasaki, K, Takagi, J. | | Deposit date: | 2016-06-29 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Conformational Freedom of the LRP6 Ectodomain Is Regulated by N-glycosylation and the Binding of the Wnt Antagonist Dkk1
Cell Rep, 18, 2017
|
|
6IKN
 
 | | Crystal structure of the GAS7 F-BAR domain | | Descriptor: | Growth arrest-specific protein 7 | | Authors: | Hanawa-Suetsugu, K, Itoh, Y, Kohda, D, Shimada, A, Suetsugu, S. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phagocytosis is mediated by two-dimensional assemblies of the F-BAR protein GAS7.
Nat Commun, 10, 2019
|
|
6IKO
 
 | | Crystal structure of mouse GAS7cb | | Descriptor: | Growth arrest-specific protein 7 | | Authors: | Hanawa-Suetsugu, K, Itoh, Y, Kohda, D, Shimada, A, Suetsugu, S. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.756 Å) | | Cite: | Phagocytosis is mediated by two-dimensional assemblies of the F-BAR protein GAS7.
Nat Commun, 10, 2019
|
|
6IOL
 
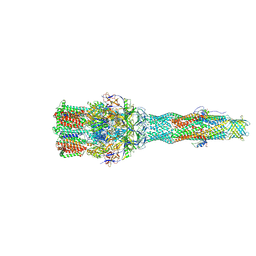 | | Cryo-EM structure of multidrug efflux pump MexAB-OprM (60 degree state) | | Descriptor: | Multidrug resistance protein MexA, Multidrug resistance protein MexB, Outer membrane protein OprM | | Authors: | Tsutsumi, K, Yonehara, R, Nakagawa, A, Yamashita, E. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structures of the wild-type MexAB-OprM tripartite pump reveal its complex formation and drug efflux mechanism.
Nat Commun, 10, 2019
|
|
6IOK
 
 | | Cryo-EM structure of multidrug efflux pump MexAB-OprM (0 degree state) | | Descriptor: | Multidrug resistance protein MexA, Multidrug resistance protein MexB, Outer membrane protein OprM | | Authors: | Tsutsumi, K, Yonehara, R, Nakagawa, A, Yamashita, E. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-03 | | Last modified: | 2019-04-17 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structures of the wild-type MexAB-OprM tripartite pump reveal its complex formation and drug efflux mechanism.
Nat Commun, 10, 2019
|
|
2ZTN
 
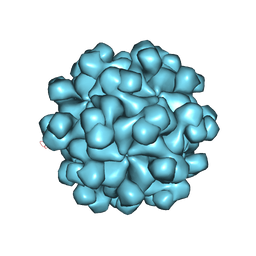 | | Hepatitis E virus ORF2 (Genotype 3) | | Descriptor: | Capsid protein | | Authors: | Yamashita, T, Unno, H, Mori, Y, Li, T.C, Takeda, N, Matsuura, Y. | | Deposit date: | 2008-10-08 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7CEA
 
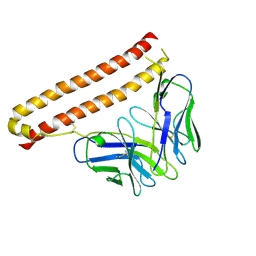 | |
7CEB
 
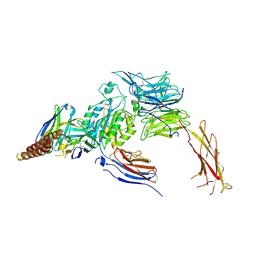 | |
5YL0
 
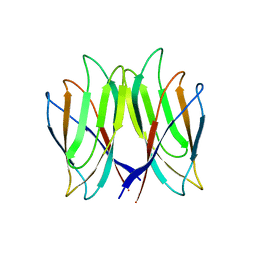 | | The crystal structure of Penaeus vannamei nodavirus P-domain (P212121) | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YKX
 
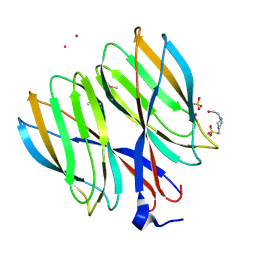 | | The crystal structure of Macrobrachium rosenbergii nodavirus P-domain with Cd ion | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, Capsid protein, ... | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
