7LPI
 
 | | APE1 phosphorothioate substrate complex with abasic ribonucleotide DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-R(P*(YA4))-D(P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker.
Comput Struct Biotechnol J, 19, 2021
|
|
6O5U
 
 | | AAC-VIa bound to Kanamycin A | | Descriptor: | Aminoglycoside N(3)-acetyltransferase, KANAMYCIN A, MAGNESIUM ION | | Authors: | Kumar, P, Cuneo, M.J. | | Deposit date: | 2019-03-04 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6NTJ
 
 | | Neutron/X-ray crystal structure of AAC-VIa bound to gentamicin C1A | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2019-01-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5EJ3
 
 | | Crystal structure of XlnB2 | | Descriptor: | Endo-1,4-beta-xylanase B | | Authors: | Couture, J.-F. | | Deposit date: | 2015-11-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.314 Å) | | Cite: | Ligand Binding Enhances Millisecond Conformational Exchange in Xylanase B2 from Streptomyces lividans.
Biochemistry, 55, 2016
|
|
5HM4
 
 | | Crystal structure of oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (TM1226) from THERMOTOGA MARITIMA at 2.0 A resolution | | Descriptor: | CALCIUM ION, Mannoside ABC transport system, sugar-binding protein | | Authors: | Lu, X, Ghimire-Rijal, S, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2016-01-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Periplasmic Binding Protein Dimer Has a Second Allosteric Event Tied to Ligand Binding.
Biochemistry, 56, 2017
|
|
6X76
 
 | | Rev1 L325G Mn2+-facilitated Product Complex with second dCTP bound | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*TP*GP*TP*GP*GP*TP*AP*GP*C)-3'), ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X72
 
 | | Rev1 Mg2+-facilitated Product Complex with two monophosphates | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*TP*GP*TP*GP*GP*TP*AP*GP*C*)-3'), DNA repair protein REV1, ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X75
 
 | | Rev1 Mn2+-facilitated Product Complex with second dCTP bound | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*TP*GP*TP*GP*GP*TP*AP*GP*C)-3'), ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X6Z
 
 | | Rev1 Ternary Complex with dCTP and Ca2+ | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X73
 
 | | Rev1 Mg2+-facilitated Product Complex with one monophosphate | | Descriptor: | AMMONIUM ION, DNA (5'-D(*CP*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*TP*GP*TP*GP*GP*TP*AP*GP*C)-3'), ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X74
 
 | | Rev1 Mg2+-facilitated Product Complex with no monophosphates | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*GP*GP*GP*TP*GP*TP*GP*GP*TP*AP*GP*C*)-3'), DNA (5'-D(P*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X71
 
 | |
6X70
 
 | | Rev1-DNA Binary Complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*TP*GP*TP*GP*GP*TP*AP*G)-3'), DNA repair protein REV1, ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X77
 
 | | Rev1 R518A Ternary Complex with dCTP and Ca2+ | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6DTR
 
 | | Apo T. maritima MalE3 | | Descriptor: | SULFATE ION, maltose-binding protein MalE3 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTU
 
 | | Maltotetraose bound T. maritima MalE1 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE1 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTT
 
 | | Apo T. maritima MalE2 | | Descriptor: | maltose-binding protein MalE2 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTS
 
 | | Maltotetraose bound T. maritima MalE2 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE2 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
5TKI
 
 | |
5TKH
 
 | | Neurospora crassa polysaccharide monooxygenase 2 ascorbate treated | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Lytic polysaccharide monooxygenase, ... | | Authors: | O'Dell, W.B, Meilleur, F. | | Deposit date: | 2016-10-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Oxygen Activation at the Active Site of a Fungal Lytic Polysaccharide Monooxygenase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5TKG
 
 | | Neurospora crassa polysaccharide monooxygenase 2 resting state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Lytic polysaccharide monooxygenase, ... | | Authors: | O'Dell, W.B, Meilleur, F. | | Deposit date: | 2016-10-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Oxygen Activation at the Active Site of a Fungal Lytic Polysaccharide Monooxygenase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6W4T
 
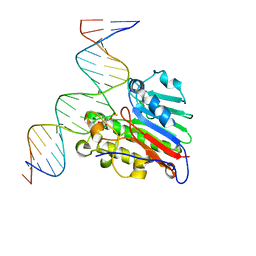 | | APE1 Y269A phosphorothioate substrate complex with abasic DNA | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*(48Z)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | AP-endonuclease 1 sculpts DNA through an anchoring tyrosine residue on the DNA intercalating loop.
Nucleic Acids Res., 48, 2020
|
|
6W4I
 
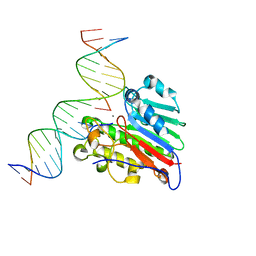 | | APE1 Y269A product complex with abasic DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(P*(3DR)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA (5'-D(P*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | AP-endonuclease 1 sculpts DNA through an anchoring tyrosine residue on the DNA intercalating loop.
Nucleic Acids Res., 48, 2020
|
|
7T5C
 
 | |
7T5E
 
 | |
