8IG8
 
 | |
8IG7
 
 | |
8IG9
 
 | |
8IGB
 
 | |
8WJO
 
 | | Cryo-EM structure of 8-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.04 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8WJL
 
 | | Cryo-EM structure of 6-subunit Smc5/6 hinge region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.15 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8WJN
 
 | | Cryo-EM structure of 6-subunit Smc5/6 head region | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosomes element 1, Non-structural maintenance of chromosomes element 4, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.58 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8IM6
 
 | | Crystal structure of HCoV 229E main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhou, Y.R, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of main proteases of HCoV-229E bound to inhibitor PF-07304814 and PF-07321332.
Biochem.Biophys.Res.Commun., 657, 2023
|
|
2I7K
 
 | | Solution Structure of the Bromodomain of Human BRD7 Protein | | Descriptor: | Bromodomain-containing protein 7 | | Authors: | Sun, H, Liu, J, Zhang, J, Huang, H, Wu, J, Shi, Y. | | Deposit date: | 2006-08-31 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4
Biochem.Biophys.Res.Commun., 358, 2007
|
|
8HZR
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, X.Y, Zhang, J, Li, J. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
S46F mutant in complex with PF07321332
To Be Published
|
|
6POM
 
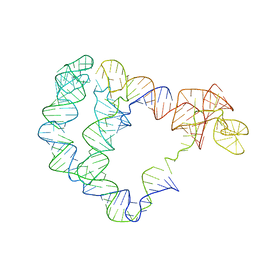 | | Cryo-EM structure of the full-length Bacillus subtilis glyQS T-box riboswitch in complex with tRNA-Gly | | Descriptor: | T-box GlyQS leader (155-MER), tRNAGly (75-MER) | | Authors: | Li, S, Su, Z, Zhang, J, Chiu, W. | | Deposit date: | 2019-07-04 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2IEL
 
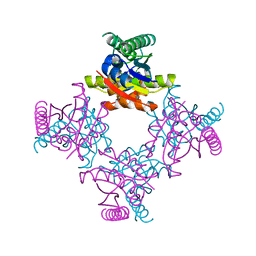 | | CRYSTAL STRUCTURE OF TT0030 from Thermus Thermophilus | | Descriptor: | Hypothetical Protein TT0030 | | Authors: | Zhu, J, Huang, J, Stepanyuk, G, Chen, L, Chang, J, Zhao, M, Xu, H, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-09-19 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF TT0030 from Thermus Thermophilus AT 1.6 ANGSTROMS RESOLUTION
To be Published
|
|
7T63
 
 | |
6VXP
 
 | |
6WA3
 
 | | Solution NMR structure of the myristoylated feline immunodeficiency virus matrix protein | | Descriptor: | MYRISTIC ACID, Matrix protein | | Authors: | Brown, J.B, Summers, H.R, Brown, L.A, Marchant, J, Canova, P.N, O'Hern, C.T, Abbott, S, Nyaunu, C, Maxwell, S, Johnson, T, Moser, M, Ablan, S.D, Carter, H, Freed, E.O, Summers, M.F. | | Deposit date: | 2020-03-24 | | Release date: | 2020-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Studies of the Rare Myristoylation Signal of the Feline Immunodeficiency Virus.
J.Mol.Biol., 432, 2020
|
|
6WA4
 
 | | Solution NMR structure of the unmyristoylated feline immunodeficiency virus matrix protein | | Descriptor: | Matrix protein | | Authors: | Brown, J.B, Summers, H.R, Brown, L.A, Marchant, J, Canova, P.N, O'Hern, C.T, Abbott, S.T, Nyaunu, C, Maxwell, S, Johnson, T, Moser, M.B, Carter, H, Ablan, S, Freed, E.O, Summers, M.F. | | Deposit date: | 2020-03-24 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Studies of the Rare Myristoylation Signal of the Feline Immunodeficiency Virus.
J.Mol.Biol., 432, 2020
|
|
6WA5
 
 | | Solution NMR Structure of the G4L/Q5K/G6S (NOS) Unmyristoylated Feline Immunodeficiency Virus Matrix Protein | | Descriptor: | Matrix protein | | Authors: | Brown, J.B, Summers, H.R, Brown, L.A, Marchant, J, Canova, P.N, O'Hern, C.T, Abbott, S.T, Nyaunu, C, Maxwell, S, Johnson, T, Moser, M.B, Ablan, S.A, Carter, H, Freed, E.O, Summers, M.F. | | Deposit date: | 2020-03-24 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Studies of the Rare Myristoylation Signal of the Feline Immunodeficiency Virus.
J.Mol.Biol., 432, 2020
|
|
5XJX
 
 | | Pre-formed plant receptor ERL1-TMM complex | | Descriptor: | LRR receptor-like serine/threonine-protein kinase ERL1, Protein TOO MANY MOUTHS | | Authors: | Chai, J, Lin, G, Zhang, L, Han, Z, Yang, X, Liu, W, Qi, Y, Chang, J, Li, E. | | Deposit date: | 2017-05-04 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.055 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
8H4Q
 
 | |
8H4H
 
 | |
4R0X
 
 | | Allosteric coupling of conformational transitions in the FK1 domain of FKBP51 near the site of steroid receptor interaction | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | LeMaster, D.M, Mustafi, S.M, Brecher, M, Zhang, J, Heroux, A, Li, H.M, Hernandez, G. | | Deposit date: | 2014-08-02 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Coupling of Conformational Transitions in the N-terminal Domain of the 51-kDa FK506-binding Protein (FKBP51) Near Its Site of Interaction with the Steroid Receptor Proteins.
J.Biol.Chem., 290, 2015
|
|
3EB7
 
 | | Crystal Structure of Insecticidal Delta-Endotoxin Cry8Ea1 from Bacillus Thuringiensis at 2.2 Angstroms Resolution | | Descriptor: | ACETATE ION, Insecticidal Delta-Endotoxin Cry8Ea1, SULFATE ION | | Authors: | Guo, S, Ye, S, Song, F, Zhang, J, Wei, L, Shu, C.L. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Bacillus thuringiensis Cry8Ea1: An insecticidal toxin toxic to underground pests, the larvae of Holotrichia parallela.
J.Struct.Biol., 168, 2009
|
|
5EJO
 
 | | Crystal structure of the winged helix domain in Chromatin assembly factor 1 subunit p90 | | Descriptor: | Chromatin assembly factor 1 subunit p90 | | Authors: | Zhang, K, Gao, Y, Li, J, Burgess, R, Han, J, Liang, H, Zhang, Z, Liu, Y. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A DNA binding winged helix domain in CAF-1 functions with PCNA to stabilize CAF-1 at replication forks
Nucleic Acids Res., 44, 2016
|
|
2ID0
 
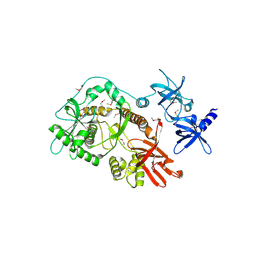 | | Escherichia coli RNase II | | Descriptor: | Exoribonuclease 2, MANGANESE (II) ION | | Authors: | Zuo, Y, Zhang, J, Wang, Y, Malhotra, A. | | Deposit date: | 2006-09-13 | | Release date: | 2006-10-03 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for Processivity and Single-Strand Specificity of RNase II.
Mol.Cell, 24, 2006
|
|
2J2F
 
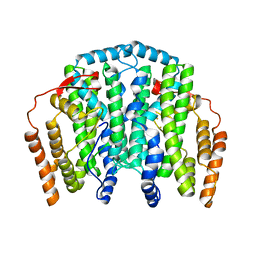 | | The T199D Mutant of Stearoyl Acyl Carrier Protein Desaturase from Ricinus Communis (Castor Bean) | | Descriptor: | ACYL-[ACYL-CARRIER-PROTEIN] DESATURASE, FE (III) ION | | Authors: | Guy, J.E, Abreu, I.A, Moche, M, Lindqvist, Y, Whittle, E, Shanklin, J. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Single Mutation in the Castor {Delta}9-18:0- Desaturase Changes Reaction Partitioning from Desaturation to Oxidase Chemistry.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
