1HCY
 
 | |
3T8X
 
 | | Crystal structure of human CD1b in complex with synthetic antigenic diacylsulfoglycolipid SGL12 and endogenous spacer | | Descriptor: | 2-O-sulfo-alpha-D-glucopyranosyl 2-O-hexadecanoyl-3-O-[(2E,4S,6S,8S)-2,4,6,8-tetramethyltetracos-2-enoyl]-alpha-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Garcia-Alles, L.F, Maveyraud, L, Mourey, L, Julien, S. | | Deposit date: | 2011-08-02 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural reorganization of the human CD1b Antigen-binding groove for presentation of mycobacterial sulfoglycolipids
To be Published
|
|
3GCM
 
 | |
2XCU
 
 | | Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP | | Descriptor: | 3-DEOXY-D-MANNO-2-OCTULOSONIC ACID TRANSFERASE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Schmidt, H, Hansen, G, Hilgenfeld, R, Mamat, U, Mesters, J.R. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and Mechanistic Analysis of the Membrane-Embedded Glycosyltransferase Waaa Required for Lipopolysaccharide Synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4RPH
 
 | |
4G1M
 
 | | Re-refinement of alpha V beta 3 structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Springer, T.A, Mi, L, Zhu, J. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Alpha V Beta 3 Integrin Crystal Structures and their Functional Implications
Biochemistry, 51, 2012
|
|
4RPJ
 
 | |
7NB4
 
 | | Structure of Mcl-1 complex with compound 1 | | Descriptor: | (2~{R})-2-[[5-(3-chloranyl-2-methyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Effect of Core Replacement on S64315, a Selective MCL-1 Inhibitor, and Its Analogues.
Acs Omega, 6, 2021
|
|
7NB7
 
 | | Structure of Mcl-1 complex with compound 6b | | Descriptor: | (2~{R})-2-[[7-but-2-ynyl-5-(3-chloranyl-2-methyl-phenyl)-6-ethyl-pyrrolo[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The Effect of Core Replacement on S64315, a Selective MCL-1 Inhibitor, and Its Analogues.
Acs Omega, 6, 2021
|
|
7JU8
 
 | | X-ray structure of MMP-13 in Complex with 4-(1,2,3-thiadiazol-4-yl)pyridine | | Descriptor: | 4-(1,2,3-thiadiazol-4-yl)pyridine, CALCIUM ION, Collagenase 3, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2020-08-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Indole Inhibitors of MMP-13 for Arthritic Disorders
Acs Omega, 6, 2021
|
|
3H1C
 
 | |
4AK9
 
 | |
4RPL
 
 | | Crystal structure of Micobacterium tuberculosis UDP-Galactopyranose mutase in complex with tetrafluorinated substrate analog UDP-F4-Galp | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-galactopyranose mutase, [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,5S,6R)-3,3,4,4-tetrafluoro-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl dihydrogen diphosphate (non-preferred name) | | Authors: | Van Straaten, K.E, Sanders, D.A.R. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2499 Å) | | Cite: | Structural Basis of Ligand Binding to UDP-Galactopyranose Mutase from Mycobacterium tuberculosis Using Substrate and Tetrafluorinated Substrate Analogues.
J.Am.Chem.Soc., 137, 2015
|
|
4RPG
 
 | |
3GLL
 
 | | Crystal structure of Polynucleotide Phosphorylase (PNPase) core | | Descriptor: | Polyribonucleotide nucleotidyltransferase | | Authors: | Nurmohamed, S, Luisi, B.L. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Escherichia coli polynucleotide phosphorylase core bound to RNase E, RNA and manganese: implications for catalytic mechanism and RNA degradosome assembly.
J.Mol.Biol., 389, 2009
|
|
4RPK
 
 | | Crystal structure of Micobacterium tuberculosis UDP-Galactopyranose mutase in complex with tetrafluorinated substrate analog UDP-F4-Galf | | Descriptor: | (2R,5S)-5-[(1R)-1,2-dihydroxyethyl]-3,3,4,4-tetrafluorotetrahydrofuran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), FLAVIN-ADENINE DINUCLEOTIDE, UDP-galactopyranose mutase | | Authors: | Van Straaten, K.E, Sanders, D.A.R. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Ligand Binding to UDP-Galactopyranose Mutase from Mycobacterium tuberculosis Using Substrate and Tetrafluorinated Substrate Analogues.
J.Am.Chem.Soc., 137, 2015
|
|
3GME
 
 | |
4I6O
 
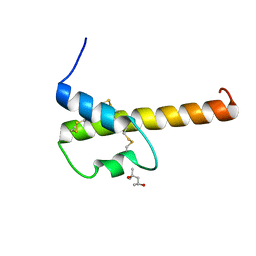 | | Crystal structure of chemically synthesized human anaphylatoxin C3a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Complement C3 | | Authors: | Wang, C.I.A, Ghassemian, A, Collins, B, Lewis, R.J, Alewood, P.F, Durek, T. | | Deposit date: | 2012-11-29 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Efficient chemical synthesis of human complement protein C3a.
Chem.Commun.(Camb.), 49, 2013
|
|
1QFF
 
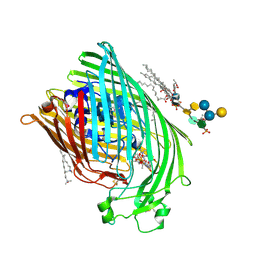 | | E. COLI FERRIC HYDROXAMATE UPTAKE RECEPTOR (FHUA) IN COMPLEX WITH BOUND FERRICHROME-IRON | | Descriptor: | 2-AMINO-VINYL-PHOSPHATE, 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Ferguson, A.D, Hofmann, E, Coulton, J.W, Diederichs, K, Welte, W. | | Deposit date: | 1999-04-10 | | Release date: | 2000-07-26 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins.
Structure Fold.Des., 8, 2000
|
|
1QD8
 
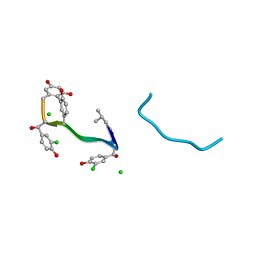 | | COMPLEX OF VANCOMYCIN WITH N-ACETYL GLYCINE | | Descriptor: | ACETYLAMINO-ACETIC ACID, CHLORIDE ION, VANCOMYCIN, ... | | Authors: | Loll, P.J, Kaplan, J, Selinsky, B, Axelsen, P.H. | | Deposit date: | 1999-07-15 | | Release date: | 1999-08-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Vancomycin Binding to Low-Affinity Ligands: Delineating a Minimum Set of Interactions Necessary for High-Affinity Binding.
J.Med.Chem., 42, 1999
|
|
1ODF
 
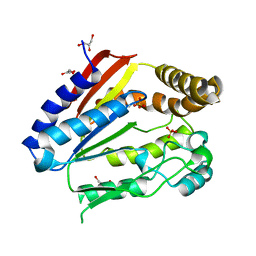 | | Structure of YGR205w protein. | | Descriptor: | GLYCEROL, HYPOTHETICAL 33.3 KDA PROTEIN IN ADE3-SER2 INTERGENIC REGION, SULFATE ION | | Authors: | Li De La Sierra-Gallay, I, Van Tilbeurgh, H. | | Deposit date: | 2003-02-19 | | Release date: | 2003-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Ygr205W Protein from Saccharomyces Cerevisiae: Close Structural Resemblance to E.Coli Pantothenate Kinase
Proteins: Struct.,Funct., Genet., 54, 2004
|
|
2BDN
 
 | | Crystal structure of human MCP-1 bound to a blocking antibody, 11K2 | | Descriptor: | Antibody heavy chain 11K2, Antibody light chain 11K2, Small inducible cytokine A2 | | Authors: | Boriack-Sjodin, P.A, Rushe, M, Reid, C, Jarpe, M, van Vlijmen, H, Bailly, V. | | Deposit date: | 2005-10-20 | | Release date: | 2006-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure activity relationships of monocyte chemoattractant proteins in complex with a blocking antibody.
Protein Eng.Des.Sel., 19, 2006
|
|
1FI1
 
 | | FhuA in complex with lipopolysaccharide and rifamycin CGP4832 | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Koedding, J, Boes, C, Walker, G, Coulton, J.W, Diederichs, K, Braun, V, Welte, W. | | Deposit date: | 2000-08-03 | | Release date: | 2001-08-29 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Active transport of an antibiotic rifamycin derivative by the outer-membrane protein FhuA.
Structure, 9, 2001
|
|
2WPD
 
 | | The Mg.ADP inhibited state of the yeast F1c10 ATP synthase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP SYNTHASE SUBUNIT 9, ... | | Authors: | Dautant, A, Velours, J, Giraud, M.-F. | | Deposit date: | 2009-08-05 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.432 Å) | | Cite: | Crystal Structure of the Mg.Adp-Inhibited State of the Yeast F1C10-ATP Synthase.
J.Biol.Chem., 285, 2010
|
|
2WYU
 
 | | High resolution structure of Thermus thermophilus enoyl-acyl carrier protein reductase apo-form | | Descriptor: | ENOYL-[ACYL CARRIER PROTEIN] REDUCTASE, GLYCEROL, SODIUM ION | | Authors: | Otero, J.M, Noel, A.J, Guardado-Calvo, P, Llamas-Saiz, A.L, van Raaij, M.J. | | Deposit date: | 2009-11-20 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structures of Thermus thermophilus enoyl-acyl carrier protein reductase in the apo form, in complex with NAD+ and in complex with NAD+ and triclosan.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 68, 2012
|
|
