8FNF
 
 | | Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNI
 
 | | Cryo-EM structure of RNase-treated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNK
 
 | | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
9IJV
 
 | | Neuraminidase of A/Switzerland/9715293/2013 H3N2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, X, Ge, J, Li, X. | | Deposit date: | 2024-06-25 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Neuraminidase of A/Switzerland/9715293/2013 H3N2
To Be Published
|
|
6OHU
 
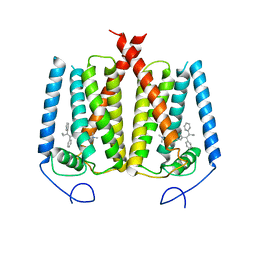 | | Structure of EBP and tamoxifen | | Descriptor: | (Z)-2-[4-(1,2)-DIPHENYL-1-BUTENYL)-PHENOXY]-N,N-DIMETHYLETHANAMINE, 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | | Authors: | Long, T, Li, X. | | Deposit date: | 2019-04-06 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.526 Å) | | Cite: | Structural basis for human sterol isomerase in cholesterol biosynthesis and multidrug recognition.
Nat Commun, 10, 2019
|
|
9EKS
 
 | | ML2-SA1 bound TRPML1-V432A/A433G in a partially open state | | Descriptor: | (3aS,4S,7R,7aS)-3-(2,6-dichlorophenyl)-3a,4,5,6,7,7a-hexahydro-4,7-methano-1,2-benzoxazole, Mucolipin-1 | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2024-12-03 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | ML2-SA1 bound TRPML1-V432A/A433G in a partially open state
To Be Published
|
|
9EL1
 
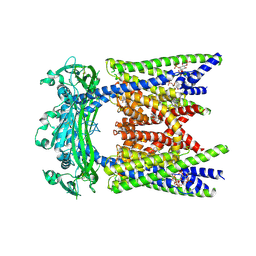 | | ML2-SA1/PI(3,5)P2 bound TRPML2 in an open state | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, (3aS,4S,7R,7aS)-3-(2,6-dichlorophenyl)-3a,4,5,6,7,7a-hexahydro-4,7-methano-1,2-benzoxazole, Mucolipin-2 | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2024-12-03 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | ML2-SA1/PI(3,5)P2 bound TRPML2 in an open state
To Be Published
|
|
9EKV
 
 | | ML-SA5 bound TRPML1 in an open state | | Descriptor: | Mucolipin-1, N4-(3-chloranyl-2-piperidin-1-yl-phenyl)-N1,N1-dimethyl-benzene-1,4-disulfonamide | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2024-12-03 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | ML-SA5 bound TRPML1 in an open state
To Be Published
|
|
9EKU
 
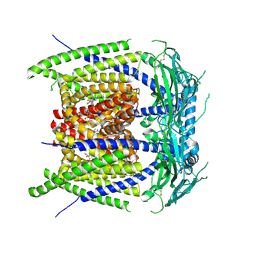 | |
9EKX
 
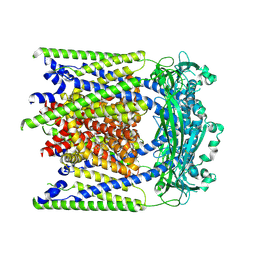 | | (-)ML-SI3 bound TRPML2 in a closed state | | Descriptor: | Mucolipin-2, N-[(1R,2R)-2-[4-(2-methoxyphenyl)piperazin-1-yl]cyclohexyl]benzenesulfonamide | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2024-12-03 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | (-)ML-SI3 bound TRPML2 in a closed state
To Be Published
|
|
9EKZ
 
 | | (+)ML-SI3 bound TRPML2 in an open state | | Descriptor: | Mucolipin-2, N-{(1S,2S)-2-[4-(2-methoxyphenyl)piperazin-1-yl]cyclohexyl}benzenesulfonamide | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2024-12-03 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | (+)ML-SI3 bound TRPML2 in an open state
To Be Published
|
|
9EKY
 
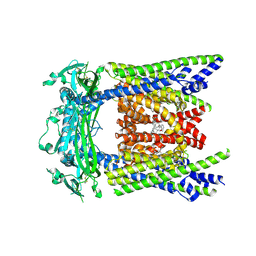 | | (+)ML-SI3 bound TRPML2 in a pre-open state | | Descriptor: | Mucolipin-2, N-{(1S,2S)-2-[4-(2-methoxyphenyl)piperazin-1-yl]cyclohexyl}benzenesulfonamide | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2024-12-03 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | (+)ML-SI3 bound TRPML2 in a pre-open state
To Be Published
|
|
9EKW
 
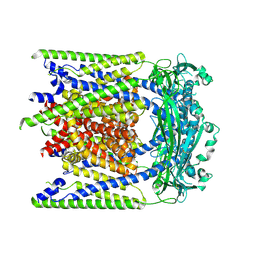 | |
9EL0
 
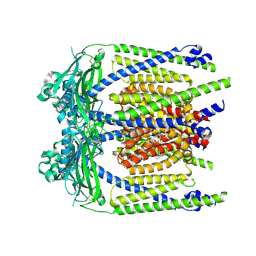 | | ML2-SA1 bound TRPML2 in a pre-open state | | Descriptor: | (3aS,4S,7R,7aS)-3-(2,6-dichlorophenyl)-3a,4,5,6,7,7a-hexahydro-4,7-methano-1,2-benzoxazole, Mucolipin-2 | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2024-12-03 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | ML2-SA1 bound TRPML2 in a pre-open state
To Be Published
|
|
9EKT
 
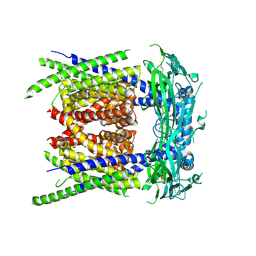 | |
9BOI
 
 | | Cryo-EM structure of human Spns1 in complex with LPC (18:1) | | Descriptor: | (4R,7R,18Z)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, mVenus,Maltose/maltodextrin-binding periplasmic protein,Protein spinster homolog 1,Designed ankyrin repeat protein (DARPin) | | Authors: | Chen, H, Li, X. | | Deposit date: | 2024-05-03 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular basis of Spns1-mediated lysophospholipid transport from the lysosome.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
7TUZ
 
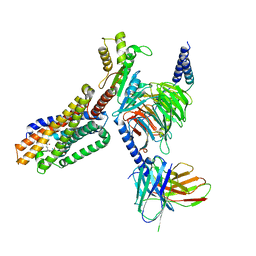 | | Cryo-EM structure of 7alpha,25-dihydroxycholesterol-bound EBI2/GPR183 in complex with Gi protein | | Descriptor: | (2S,4aS,4bS,7R,8S,8aS,9R,10aR)-7-[(2R,3R)-7-hydroxy-3,7-dimethyloctan-2-yl]-4a,7,8-trimethyltetradecahydrophenanthrene-2,9-diol, G-protein coupled receptor 183, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, H, Hung, W, Li, X. | | Deposit date: | 2022-02-03 | | Release date: | 2022-04-13 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structures of oxysterol sensor EBI2/GPR183, a key regulator of the immune response.
Structure, 30, 2022
|
|
7TUY
 
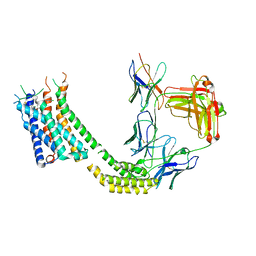 | | Cryo-EM structure of GSK682753A-bound EBI2/GPR183 | | Descriptor: | 8-[(2E)-3-(4-chlorophenyl)prop-2-enoyl]-3-[(3,4-dichlorophenyl)methyl]-1-oxa-3,8-diazaspiro[4.5]decan-2-one, G-protein coupled receptor 183,Soluble cytochrome b562 fusion, anti-BRIL Fab Heavy chain, ... | | Authors: | Chen, H, Huang, W, Li, X. | | Deposit date: | 2022-02-03 | | Release date: | 2022-04-13 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of oxysterol sensor EBI2/GPR183, a key regulator of the immune response.
Structure, 30, 2022
|
|
6LTP
 
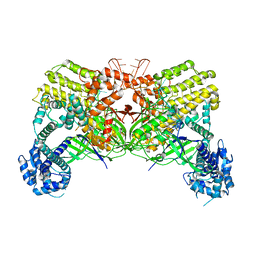 | | Crystal structure of Cas12i2 binary complex | | Descriptor: | Cas12i2, crRNA (56-mer RNA) | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
7UNE
 
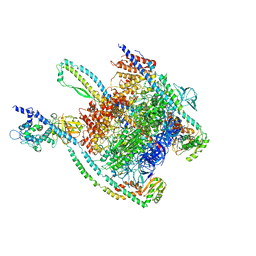 | |
7UNF
 
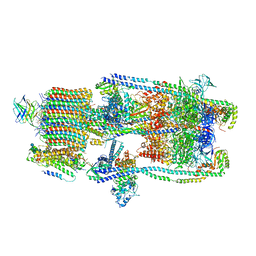 | | CryoEM structure of a mEAK7 bound human V-ATPase complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2022-04-10 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Molecular basis of mEAK7-mediated human V-ATPase regulation.
Nat Commun, 13, 2022
|
|
2OQV
 
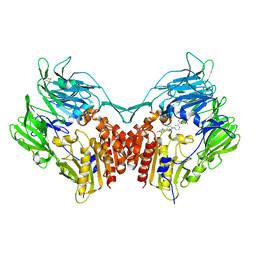 | | Human Dipeptidyl Peptidase IV (DPP4) with piperidine-constrained phenethylamine | | Descriptor: | (3R,4R)-1-{6-[3-(METHYLSULFONYL)PHENYL]PYRIMIDIN-4-YL}-4-(2,4,5-TRIFLUOROPHENYL)PIPERIDIN-3-AMINE, Dipeptidyl peptidase 4 (Dipeptidyl peptidase IV) (DPP IV) | | Authors: | Pei, Z, Li, X, von Geldern, T.W, Longenecker, K.L, Pireh, D, Stewart, K.D. | | Deposit date: | 2007-02-01 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Structure-Activity Relationships of Piperidinone- and Piperidine-Constrained Phenethylamines as Novel, Potent, and Selective Dipeptidyl Peptidase IV Inhibitors.
J.Med.Chem., 50, 2007
|
|
4QPG
 
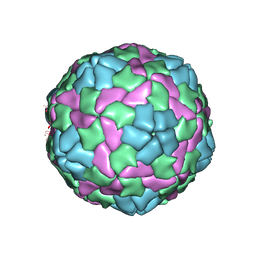 | | Crystal structure of empty hepatitis A virus | | Descriptor: | CHLORIDE ION, Capsid protein VP0, Capsid protein VP1, ... | | Authors: | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
4JGZ
 
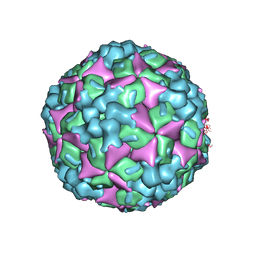 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group I222) | | Descriptor: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, McAuley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
4JGY
 
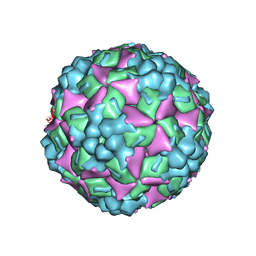 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group P4232) | | Descriptor: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, Mcauley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
