5E5K
 
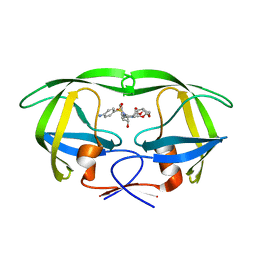 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 4.3 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 protease | | Authors: | Kovalevsky, A.Y, Das, A. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.75 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2OG8
 
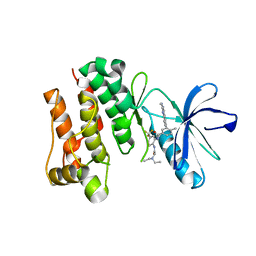 | | crystal structure of aminoquinazoline 36 bound to Lck | | Descriptor: | N-{2-[(N,N-DIETHYLGLYCYL)AMINO]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[2-(METHYLAMINO)QUINAZOLIN-6-YL]BENZAMIDE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Aminoquinazolines as Potent, Orally Bioavailable Inhibitors of Lck: Synthesis, SAR, and in Vivo Anti-Inflammatory Activity
J.Med.Chem., 49, 2006
|
|
2NTC
 
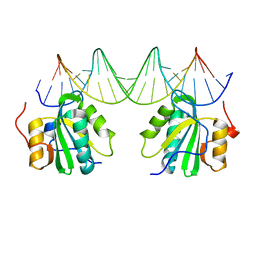 | |
6WBS
 
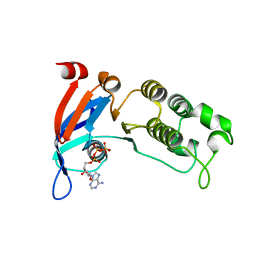 | | Human CFTR first nucleotide binding domain with dF508/V510D | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Simon, K.S, Kothe, M, Hilbert, B, Batchelor, J.D, Hurlbut, G.D. | | Deposit date: | 2020-03-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Determining the Molecular Mechanism of Suppressor Mutation V510D and the Contribution of Helical Unraveling to the dF508-CFTR Defect
To Be Published
|
|
6TH0
 
 | | Crystal structure of Arabidopsis thaliana NAA60 in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Acyl-CoA N-acyltransferases (NAT) superfamily protein | | Authors: | Layer, D, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Arabidopsis N alpha -acetyltransferase NAA60 locates to the plasma membrane and is vital for the high salt stress response.
New Phytol., 228, 2020
|
|
6TI2
 
 | |
7TUR
 
 | | Joint X-ray/neutron structure of aspastate aminotransferase (AAT) in complex with pyridoxamine 5'-phosphate (PMP) | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase, ... | | Authors: | Drago, V.N, Kovalevsky, A.Y, Dajnowicz, S, Mueser, T.C. | | Deposit date: | 2022-02-03 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | An N⋯H⋯N low-barrier hydrogen bond preorganizes the catalytic site of aspartate aminotransferase to facilitate the second half-reaction.
Chem Sci, 13, 2022
|
|
4NE9
 
 | | PCSK9 in complex with LDLR peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Low-density lipoprotein receptor, ... | | Authors: | Liu, S. | | Deposit date: | 2013-10-28 | | Release date: | 2014-09-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design and synthesis of truncated EGF-A peptides that restore LDL-R recycling in the presence of PCSK9 in vitro.
Chem.Biol., 21, 2014
|
|
2Q98
 
 | |
2GLK
 
 | | High-resolution study of D-Xylose isomerase, 0.94A resolution. | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Xylose isomerase | | Authors: | Katz, A.K, Carrell, H.L, Hanson, B.L, Harp, J.M, Glusker, J.P, Bunick, G.J. | | Deposit date: | 2006-04-05 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Locating active-site hydrogen atoms in D-xylose isomerase: Time-of-flight neutron diffraction.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DPG
 
 | | COMPLEX OF INACTIVE MUTANT (H240->N) OF GLUCOSE 6-PHOSPHATE DEHYDROGENASE FROM LEUCONOSTOC MESENTEROIDES WITH NADP+ | | Descriptor: | GLUCOSE 6-PHOSPHATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Adams, M.J, Naylor, C.E, Paludin, S, Gover, S. | | Deposit date: | 1998-04-17 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | On the mechanism of the reaction catalyzed by glucose 6-phosphate dehydrogenase.
Biochemistry, 37, 1998
|
|
1F76
 
 | | ESCHERICHIA COLI DIHYDROOROTATE DEHYDROGENASE | | Descriptor: | Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, FORMIC ACID, ... | | Authors: | Norager, S, Jensen, K.F, Bjornberg, O, Larsen, S. | | Deposit date: | 2000-06-26 | | Release date: | 2002-10-16 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E. coli Dihydroorotate Dehydrogenase Reveals Structural and Functional Distinction between different classes of
dihydroorotate dehydrogenases
Structure, 10, 2002
|
|
3EPG
 
 | | Structure of Human DNA Polymerase Iota complexed with N2-ethylguanine | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*DTP*DCP*DTP*(2EG)P*DGP*DGP*DGP*DTP*DCP*DCP*DTP*DAP*DGP*DGP*DAP*DCP*DCP*(DOC))-3', DNA polymerase iota, ... | | Authors: | Pence, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lesion bypass of N2-ethylguanine by human DNA polymerase iota.
J.Biol.Chem., 284, 2009
|
|
6EGU
 
 | |
6RX1
 
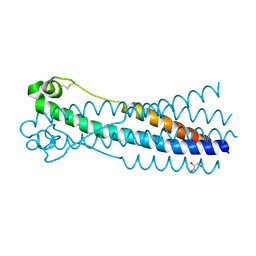 | | Crystal structure of human syncytin 1 in post-fusion conformation | | Descriptor: | CHLORIDE ION, GLYCEROL, Syncytin-1 | | Authors: | Ruigrok, K, Backovic, M, Vaney, M.C, Rey, F.A. | | Deposit date: | 2019-06-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structures of the Post-fusion 6-Helix Bundle of the Human Syncytins and their Functional Implications.
J.Mol.Biol., 431, 2019
|
|
5LK0
 
 | |
6RX3
 
 | | Crystal structure of human syncytin 2 in post-fusion conformation | | Descriptor: | CHLORIDE ION, Syncytin-2 | | Authors: | Ruigrok, K, Backovic, M, Vaney, M.C, Rey, F.A. | | Deposit date: | 2019-06-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Structures of the Post-fusion 6-Helix Bundle of the Human Syncytins and their Functional Implications.
J.Mol.Biol., 431, 2019
|
|
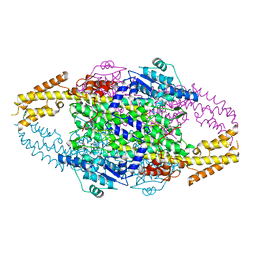
6RGS
 
 | |
5LK1
 
 | |
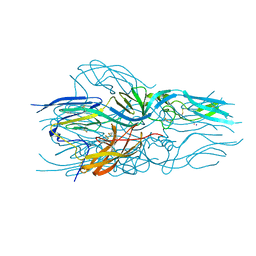
5LJX
 
 | |
6EGT
 
 | |
5LJZ
 
 | | Structure of hantavirus envelope glycoprotein Gc in postfusion conformation | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, ... | | Authors: | Guardado-Calvo, P, Rey, F.A. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanistic Insight into Bunyavirus-Induced Membrane Fusion from Structure-Function Analyses of the Hantavirus Envelope Glycoprotein Gc.
Plos Pathog., 12, 2016
|
|
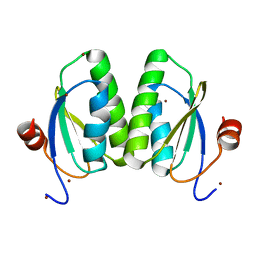
6SCP
 
 | |
5LJY
 
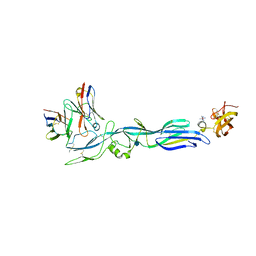 | | Structure of hantavirus envelope glycoprotein Gc in complex with scFv A5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT HEXAMMINE(III), Envelopment polyprotein, ... | | Authors: | Guardado-Calvo, P, Stettner, E, Jeffers, S.A, Rey, F.A. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanistic Insight into Bunyavirus-Induced Membrane Fusion from Structure-Function Analyses of the Hantavirus Envelope Glycoprotein Gc.
Plos Pathog., 12, 2016
|
|
5LK3
 
 | |
