7LAZ
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD3 bound to ERK5-IN-1 | | Descriptor: | 11-cyclopentyl-2-({2-ethoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 3 | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2021-01-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LB2
 
 | | Crystal structure of the tandem bromodomain (BD1, BD2) of human TAF1 bound to ZS1-589 | | Descriptor: | (3R)-3-methyl-4-{6-[1-(S-methylsulfonimidoyl)cyclopropyl]-2-(naphthalen-1-yl)pyrimidin-4-yl}morpholine, 1,2-ETHANEDIOL, HEXANE-1,6-DIOL, ... | | Authors: | Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the tandem bromodomain (BD1, BD2) of human TAF1 bound to ZS1-589
To Be Published
|
|
7LEJ
 
 | | Crystal structure of the second bromodomain (BD2) of human BRDT bound to Volasertib | | Descriptor: | Bromodomain testis-specific protein, N-{trans-4-[4-(cyclopropylmethyl)piperazin-1-yl]cyclohexyl}-4-{[(7R)-7-ethyl-5-methyl-8-(1-methylethyl)-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxybenzamide | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LB0
 
 | |
7LBT
 
 | | Crystal structure of the second bromodomain (BD2) of human BRD3 bound to ERK5-IN-1 | | Descriptor: | 11-cyclopentyl-2-({2-ethoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 3 | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2021-01-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LB1
 
 | |
7LEL
 
 | |
7LAY
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD3 bound to SG3-179 | | Descriptor: | 1,2-ETHANEDIOL, 4-[[4-[[3-(~{tert}-butylsulfonylamino)-4-chloranyl-phenyl]amino]-5-methyl-pyrimidin-2-yl]amino]-2-fluoranyl-~{N}-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 3, ... | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2021-01-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LEK
 
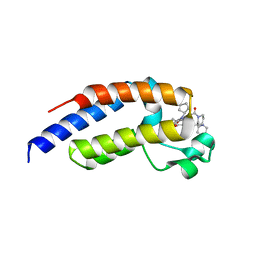 | | Crystal structure of the second bromodomain (BD2) of human BRDT bound to ERK5-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 11-cyclopentyl-2-({2-ethoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain testis-specific protein, ... | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LAK
 
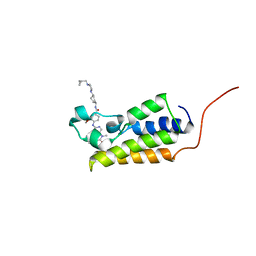 | | Crystal structure of the first bromodomain (BD1) of human BRD2 bound to volasertib | | Descriptor: | Bromodomain-containing protein 2, N-{trans-4-[4-(cyclopropylmethyl)piperazin-1-yl]cyclohexyl}-4-{[(7R)-7-ethyl-5-methyl-8-(1-methylethyl)-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxybenzamide | | Authors: | Karim, M.R, Bikowitz, M, Chan, A, Schonbrunn, E. | | Deposit date: | 2021-01-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LEM
 
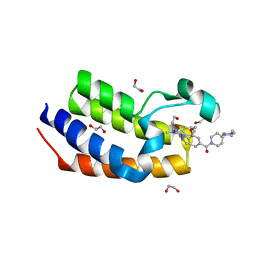 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to LRRK2-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain testis-specific protein | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7KDZ
 
 | |
1YBG
 
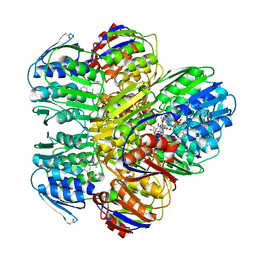 | | MurA inhibited by a derivative of 5-sulfonoxy-anthranilic acid | | Descriptor: | N-METHYL-N-{2-[(2-NAPHTHYLSULFONYL)AMINO]-5-[(2-NAPHTHYLSULFONYL)OXY]BENZOYL}-L-ASPARTIC ACID, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Eschenburg, S, Priestman, M.A, Abdul-Latif, F.A, Delachaume, C, Fassy, F, Schonbrunn, E. | | Deposit date: | 2004-12-20 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel Inhibitor That Suspends the Induced Fit Mechanism of UDP-N-acetylglucosamine Enolpyruvyl Transferase (MurA).
J.Biol.Chem., 280, 2005
|
|
7LRK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-093 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{S})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-16 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LP0
 
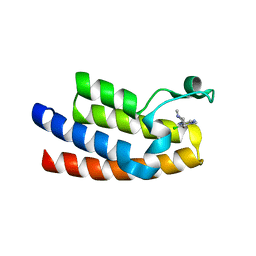 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-077 | | Descriptor: | 1,2-ETHANEDIOL, 4-chlorol-2-methyl-5-[[(3~{R})-1-methylpiperidin-3-yl]amino]pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LRO
 
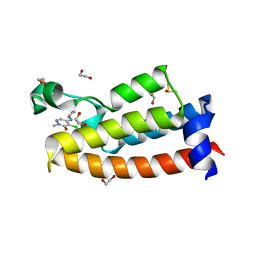 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-01-105 | | Descriptor: | 1,2-ETHANEDIOL, 5-(azetidin-3-ylamino)-4-chloranyl-2-methyl-pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LPK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-03-112 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{R})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-12 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
1ZCM
 
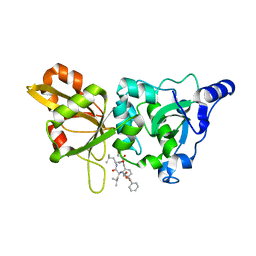 | | Human calpain protease core inhibited by ZLLYCH2F | | Descriptor: | CALCIUM ION, Calpain 1, large [catalytic] subunit, ... | | Authors: | Li, Q, Hanzlik, R.P, Weaver, R.F, Schonbrunn, E. | | Deposit date: | 2005-04-12 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mode of action of a covalently inhibiting peptidomimetic on the human calpain protease core
Biochemistry, 45, 2006
|
|
2AA9
 
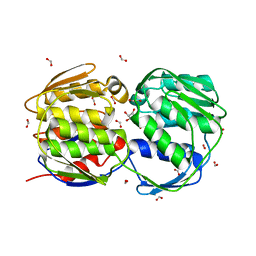 | | EPSP synthase liganded with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID | | Authors: | Priestman, M.A, Healy, M.L, Funke, T, Becker, A, Schonbrunn, E. | | Deposit date: | 2005-07-13 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis for the glyphosate-insensitivity of the reaction of 5-enolpyruvylshikimate 3-phosphate synthase with shikimate.
Febs Lett., 579, 2005
|
|
1RYW
 
 | | C115S MurA liganded with reaction products | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2003-12-22 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence That the Fosfomycin Target Cys115 in UDP-N-acetylglucosamine Enolpyruvyl Transferase (MurA) Is Essential for Product Release.
J.Biol.Chem., 280, 2005
|
|
1Q36
 
 | | EPSP synthase (Asp313Ala) liganded with tetrahedral reaction intermediate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, 5-(1-CARBOXY-1-PHOSPHONOOXY-ETHOXYL)-4-HYDROXY-3-PHOSPHONOOXY-CYCLOHEX-1-ENECARBOXYLIC ACID, FORMIC ACID | | Authors: | Eschenburg, S, Kabsch, W, Healy, M.L, Schonbrunn, E. | | Deposit date: | 2003-07-28 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A New View of the Mechanisms of UDP-N-Acetylglucosamine Enolpyruvyl Transferase (MurA) and 5-Enolpyruvylshikimate-3-phosphate Synthase (AroA) Derived from X-ray Structures of Their Tetrahedral Reaction Intermediate States.
J.Biol.Chem., 278, 2003
|
|
1Q3G
 
 | | MurA (Asp305Ala) liganded with tetrahedral reaction intermediate | | Descriptor: | 1,2-ETHANEDIOL, 3'-1-CARBOXY-1-PHOSPHONOOXY-ETHOXY-URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Eschenburg, S, Kabsch, W, Healy, M.L, Schonbrunn, E. | | Deposit date: | 2003-07-29 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A New View of the Mechanisms of UDP-N-Acetylglucosamine Enolpyruvyl Transferase (MurA) and 5-Enolpyruvylshikimate-3-phosphate Synthase (AroA) Derived from X-ray Structures of Their Tetrahedral Reaction Intermediate States.
J.Biol.Chem., 278, 2003
|
|
8U4E
 
 | | Cryo-EM structure of long form insulin receptor (IR-B) with three IGF2 bound, asymmetric conformation. | | Descriptor: | Insulin receptor, Insulin-like growth factor II | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8U4B
 
 | | Cryo-EM structure of long form insulin receptor (IR-B) in the apo state | | Descriptor: | Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8U4C
 
 | | Cryo-EM structure of long form insulin receptor (IR-B) with four IGF2 bound, symmetric conformation. | | Descriptor: | Insulin receptor, Insulin-like growth factor II | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
