7PK3
 
 | | Notum_ARUK3001185 | | Descriptor: | 1-[2,4-bis(chloranyl)-3-(trifluoromethyl)phenyl]-1,2,3-triazole, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Vecchia, L, Hillier, J, Zhao, Y, Fish, P, Jones, E.Y. | | Deposit date: | 2021-08-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Design of a Potent, Selective, and Brain-Penetrant Inhibitor of Wnt-Deactivating Enzyme Notum by Optimization of a Crystallographic Fragment Hit.
J.Med.Chem., 65, 2022
|
|
5K2C
 
 | | 1.9 angstrom A2a adenosine receptor structure with sulfur SAD phasing and phase extension using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
7PKV
 
 | | Notum_Inhibitor ARUK3000223 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Ruza, R, Zhao, Y, Fish, P, Jones, E.Y. | | Deposit date: | 2021-08-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Design of a Potent, Selective, and Brain-Penetrant Inhibitor of Wnt-Deactivating Enzyme Notum by Optimization of a Crystallographic Fragment Hit.
J.Med.Chem., 65, 2022
|
|
5K2A
 
 | | 2.5 angstrom A2a adenosine receptor structure with sulfur SAD phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5KCU
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, alpha-naphthyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(naphthalen-2-yl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCF
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, 4-methoxybenzyl OBHS-N derivative | | Descriptor: | (1R,2S,4R)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, ... | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCT
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, 4-chlorobenzyl OBHS-N derivative | | Descriptor: | (1R,2S,4R)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, ... | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCD
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-methyl Substituted OBHS-N derivative | | Descriptor: | (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-N-methyl-N-phenyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KQA
 
 | | Crystal structure of buckwheat glutaredoxin-glutathione complex | | Descriptor: | GLUTATHIONE, Glutaredoxin-glutathione complex | | Authors: | Zhang, X, Wang, W, Zhao, Y, Wang, Z, Wang, H. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the binding of buckwheat glutaredoxin with GSH and regulation of its catalytic activity
J. Inorg. Biochem., 173, 2017
|
|
4N92
 
 | | Crystal structure of beta-lactamse PenP_E166S | | Descriptor: | Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-19 | | Release date: | 2014-09-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
4OCH
 
 | | Apo structure of Smr domain of MutS2 from Deinococcus radiodurans | | Descriptor: | Endonuclease MutS2, GLYCEROL | | Authors: | Zhang, H, Zhao, Y, Xu, Q, Hua, Y.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4001 Å) | | Cite: | Structural and functional studies of MutS2 from Deinococcus radiodurans.
Dna Repair, 21, 2014
|
|
6ZCZ
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in ternary complex with EY6A Fab and a nanobody. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EY6A heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5VOT
 
 | | Structure of AMPA receptor-TARP complex | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Chen, S, Zhao, Y, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
4OD6
 
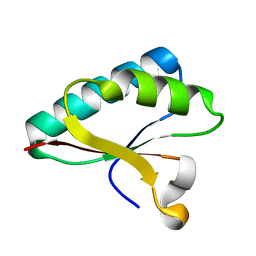 | | Structure of Smr domain of MutS2 from Deinococcus radiodurans, Mn2+ soaked | | Descriptor: | Endonuclease MutS2 | | Authors: | Zhang, H, Zhao, Y, Xu, Q, Hua, Y.J. | | Deposit date: | 2014-01-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Structural and functional studies of MutS2 from Deinococcus radiodurans.
Dna Repair, 21, 2014
|
|
5VOU
 
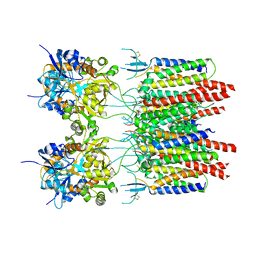 | | Structure of AMPA receptor-TARP complex | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Chen, S, Zhao, Y, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
4N9L
 
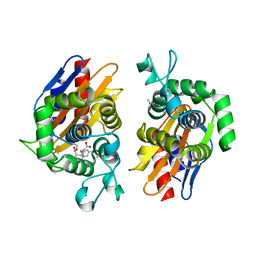 | | crystal structure of beta-lactamse PenP_E166S in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
4N9K
 
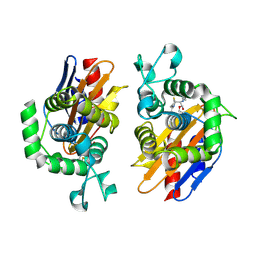 | | crystal structure of beta-lactamse PenP_E166S in complex with cephaloridine | | Descriptor: | 5-METHYL-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
3JWP
 
 | | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, TRIETHYLENE GLYCOL, Transcriptional regulatory protein sir2 homologue, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Lin, Y.H, MacKenzie, F, Senisterra, G, Allali-Hassanali, A, Vedadi, M, Ravichandran, M, Cossar, D, Kozieradzki, I, Zhao, Y, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Qiu, W, Brand, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-18 | | Release date: | 2009-10-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP
TO BE PUBLISHED
|
|
1F0J
 
 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 4B2B | | Descriptor: | ARSENIC, MAGNESIUM ION, PHOSPHODIESTERASE 4B, ... | | Authors: | Xu, R.X, Hassell, A.M, Vanderwall, D, Lambert, M.H, Holmes, W.D, Luther, M.A, Rocque, W.J, Milburn, M.V, Zhao, Y, Ke, H, Nolte, R.T. | | Deposit date: | 2000-05-16 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity.
Science, 288, 2000
|
|
6PNY
 
 | | X-ray Structure of Flpp3 | | Descriptor: | Flpp3 | | Authors: | Zook, J.D, Shekhar, M, Hansen, D.T, Conrad, C, Grant, T.D, Gupta, C, White, T, Barty, A, Basu, S, Zhao, Y, Zatsepin, N.A, Ishchenko, A, Batyuk, A, Gati, C, Li, C, Galli, L, Coe, J, Hunter, M, Liang, M, Weierstall, U, Nelson, G, James, D, Stauch, B, Craciunescu, F, Thifault, D, Liu, W, Cherezov, V, Singharoy, A, Fromme, P. | | Deposit date: | 2019-07-03 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | XFEL and NMR Structures of Francisella Lipoprotein Reveal Conformational Space of Drug Target against Tularemia.
Structure, 28, 2020
|
|
4ZEL
 
 | | Human dopamine beta-hydroxylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Vendelboe, T.V, Harris, P, Christensen, H.E.M, Harlos, K, Walter, T, Zhao, Y, Omari, K. | | Deposit date: | 2015-04-20 | | Release date: | 2016-04-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of human dopamine beta-hydroxylase at 2.9 angstrom resolution.
Sci Adv, 2, 2016
|
|
4Z7I
 
 | | Crystal structure of insulin regulated aminopeptidase in complex with ligand | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DG025 transition-state analogue enzyme inhibitor, ... | | Authors: | Mpakali, A, Saridakis, E, Harlos, K, Zhao, Y, Stratikos, E. | | Deposit date: | 2015-04-07 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal Structure of Insulin-Regulated Aminopeptidase with Bound Substrate Analogue Provides Insight on Antigenic Epitope Precursor Recognition and Processing.
J Immunol., 195, 2015
|
|
3MR2
 
 | | Human DNA polymerase eta in complex with normal DNA and incoming nucleotide (Nrm) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*T*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
3MR5
 
 | | Human DNA polymerase eta - DNA ternary complex with a CPD 1bp upstream of the active site (TT3) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*CP*GP*TP*CP*AP*TP*AP*A)-3'), DNA (5'-D(*TP*AP*AP*CP*(TTD)P*AP*TP*GP*AP*CP*GP*A)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
3MR3
 
 | | Human DNA polymerase eta - DNA ternary complex with the 3'T of a CPD in the active site (TT1) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*CP*AP*(TTD)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
