5WVN
 
 | | Crystal structure of MBS-BaeS fusion protein | | Descriptor: | Maltose-binding periplasmic protein,Two-component system sensor kinase, SULFATE ION | | Authors: | Wang, W, Zhang, Y, Ran, T, Xu, D. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Proteins, 85, 2017
|
|
8VQ4
 
 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-125A. | | Descriptor: | (8R)-6-(1-benzyl-1H-pyrazole-4-carbonyl)-N-[(2S,3R)-3-(2-cyclohexylethoxy)-1-(methylamino)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expanding the ligandable proteome by paralog hopping with covalent probes.
Biorxiv, 2024
|
|
8VQ3
 
 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-198. | | Descriptor: | (8R)-N-[(2S,3R)-3-(cyclohexylmethoxy)-1-(morpholin-4-yl)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-6-(1,3-thiazole-5-carbonyl)-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Expanding the ligandable proteome by paralog hopping with covalent probes.
Biorxiv, 2024
|
|
3PCU
 
 | | Crystal structure of human retinoic X receptor alpha ligand-binding domain complexed with LX0278 and SRC1 peptide | | Descriptor: | 2-[(2S)-6-(2-methylbut-3-en-2-yl)-7-oxo-2,3-dihydro-7H-furo[3,2-g]chromen-2-yl]propan-2-yl acetate, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Zhang, H, Zhang, Y, Shen, H, Chen, J, Li, C, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-10-22 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (+)-Rutamarin as a Dual Inducer of Both GLUT4 Translocation and Expression Efficiently Ameliorates Glucose Homeostasis in Insulin-Resistant Mice.
Plos One, 7, 2012
|
|
3UAT
 
 | |
3NTP
 
 | | Human Pin1 complexed with reduced amide inhibitor | | Descriptor: | (2R)-2-(acetylamino)-3-[(2S)-2-{[2-(1H-indol-3-yl)ethyl]carbamoyl}pyrrolidin-1-yl]propyl dihydrogen phosphate, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Zhang, Y. | | Deposit date: | 2010-07-05 | | Release date: | 2012-01-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | A reduced-amide inhibitor of Pin1 binds in a conformation resembling a twisted-amide transition state.
Biochemistry, 50, 2011
|
|
7XED
 
 | | Crystal Structure of OsCIE1-Ubox and OsUBC8 complex | | Descriptor: | U-box domain-containing protein 12, UBC core domain-containing protein | | Authors: | Zhang, Y, Yu, C.Z. | | Deposit date: | 2022-03-30 | | Release date: | 2023-10-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Release of a ubiquitin brake activates OsCERK1-triggered immunity in rice.
Nature, 629, 2024
|
|
3PWR
 
 | | HIV-1 Protease Mutant L76V complexed with Saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhang, Y, Weber, I.T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
3LZD
 
 | | Crystal structure of Dph2 from Pyrococcus horikoshii with 4Fe-4S cluster | | Descriptor: | Dph2, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Torelli, A.T, Zhang, Y, Zhu, X, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
2N78
 
 | | NMR structure of IF1 from Pseudomonas aeruginosa | | Descriptor: | Translation initiation factor IF-1 | | Authors: | Zhang, Y. | | Deposit date: | 2015-09-04 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | (1)H, (13)C and (15)N resonance assignments and secondary structure analysis of translation initiation factor 1 from Pseudomonas aeruginosa.
Biomol.Nmr Assign., 10, 2016
|
|
3PWM
 
 | | HIV-1 Protease Mutant L76V with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhang, Y, Weber, I.T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
4F6V
 
 | | Crystal structure of dehydrosqualene synthase (crtm) from s. aureus complexed with bph-1034, mg2+ and fmp. | | Descriptor: | (2E,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl dihydrogen phosphate, 2,4-dioxo-4-{[3-(3-phenoxyphenyl)propyl]amino}butanoic acid, Dehydrosqualene synthase, ... | | Authors: | Lin, F.-Y, Zhang, Y, Liu, Y.-L, Oldfield, E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.
ACS Med Chem Lett, 3, 2012
|
|
4F6X
 
 | | Crystal structure of dehydrosqualene synthase (crtm) from s. aureus complexed with bph-1112 | | Descriptor: | 1-[3-(hexyloxy)benzyl]-4-hydroxy-2-oxo-1,2-dihydropyridine-3-carboxylic acid, D(-)-TARTARIC ACID, Dehydrosqualene synthase, ... | | Authors: | Lin, F.-Y, Zhang, Y, Liu, Y.-L, Oldfield, E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.
ACS Med Chem Lett, 3, 2012
|
|
3RY0
 
 | | Crystal structure of TomN, a 4-Oxalocrotonate Tautomerase homologue in Tomaymycin biosynthetic pathway | | Descriptor: | Putative tautomerase | | Authors: | Zhang, Y, Yan, W.P, Li, W.Z, Whitman, C.P. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Kinetic, Crystallographic, and Mechanistic Characterization of TomN: Elucidation of a Function for a 4-Oxalocrotonate Tautomerase Homologue in the Tomaymycin Biosynthetic Pathway.
Biochemistry, 50, 2011
|
|
4HVS
 
 | |
8GUG
 
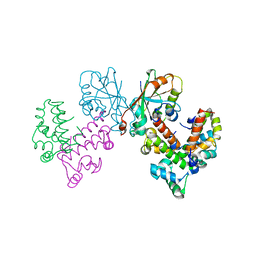 | | Structure of VPA0770 toxin bound to VPA0769 antitoxin in Vibrio parahaemolyticus | | Descriptor: | DUF2384 domain-containing protein, RES domain-containing protein | | Authors: | Song, X.J, Zhang, Y, Xu, Y.Y, Lin, Z. | | Deposit date: | 2022-09-12 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights of the toxin-antitoxin system VPA0770-VPA0769 in Vibrio parahaemolyticus.
Int.J.Biol.Macromol., 242, 2023
|
|
2M09
 
 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | Descriptor: | Splicing factor 1 | | Authors: | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
2M0G
 
 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | Descriptor: | Splicing factor 1, Splicing factor U2AF 65 kDa subunit | | Authors: | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | Deposit date: | 2012-10-25 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
5Y4O
 
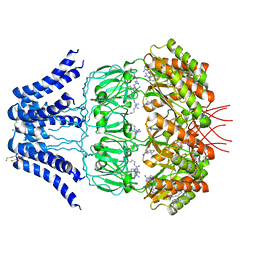 | | Cryo-EM structure of MscS channel, YnaI | | Descriptor: | Low conductance mechanosensitive channel YnaI | | Authors: | Zhang, Y, Yu, J. | | Deposit date: | 2017-08-04 | | Release date: | 2019-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A binding-block ion selective mechanism revealed by a Na/K selective channel.
Protein Cell, 9, 2018
|
|
1R0V
 
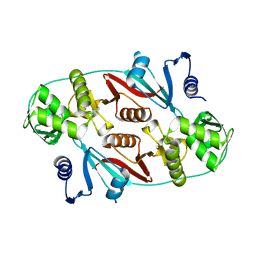 | |
1R11
 
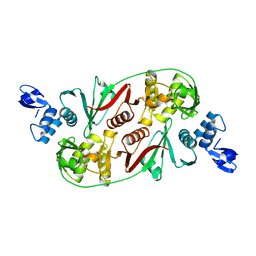 | |
4GA3
 
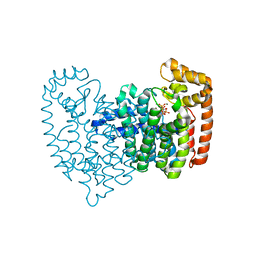 | | Crystal Structure of Human Farnesyl Diphosphate Synthase in Complex with BPH-1260 | | Descriptor: | 1-butyl-3-(2-hydroxy-2,2-diphosphonoethyl)-1H-imidazol-3-ium, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Liu, Y.-L, Zhang, Y, Oldfield, E. | | Deposit date: | 2012-07-24 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Chemo-Immunotherapeutic Anti-Malarials Targeting Isoprenoid Biosynthesis.
ACS MED.CHEM.LETT., 4, 2013
|
|
4PGV
 
 | |
8H7G
 
 | | Cryo-EM structure of the human SAGA complex | | Descriptor: | Ataxin-7, STAGA complex 65 subunit gamma, Splicing factor 3B subunit 3, ... | | Authors: | Huang, J, Zhang, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of human SAGA transcriptional coactivator complex.
Cell Discov, 8, 2022
|
|
6KQE
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-OH RNA of 4 nt | | Descriptor: | DNA (5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G*)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-08-17 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
