3PYA
 
 | |
3Q9E
 
 | | Crystal structure of H159A APAH complexed with acetylspermine | | Descriptor: | Acetylpolyamine amidohydrolase, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, POTASSIUM ION, ... | | Authors: | Lombardi, P.M, Christianson, D.W. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of prokaryotic polyamine deacetylase reveals evolutionary functional relationships with eukaryotic histone deacetylases .
Biochemistry, 50, 2011
|
|
3Q9B
 
 | | Crystal Structure of APAH complexed with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Acetylpolyamine amidohydrolase, DIMETHYL SULFOXIDE, ... | | Authors: | Lombardi, P.M, Christianson, D.W. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of prokaryotic polyamine deacetylase reveals evolutionary functional relationships with eukaryotic histone deacetylases .
Biochemistry, 50, 2011
|
|
3Q9F
 
 | | Crystal Structure of APAH complexed with CAPS | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Acetylpolyamine amidohydrolase, PHOSPHATE ION, ... | | Authors: | Lombardi, P.M, Christianson, D.W. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of prokaryotic polyamine deacetylase reveals evolutionary functional relationships with eukaryotic histone deacetylases .
Biochemistry, 50, 2011
|
|
1OKL
 
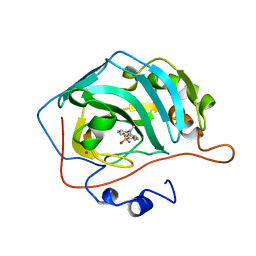 | | CARBONIC ANHYDRASE II COMPLEX WITH THE 1OKL INHIBITOR 5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONAMIDE | | Descriptor: | 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Elbaum, D, Christianson, D.W. | | Deposit date: | 1996-06-25 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected binding mode of the sulfonamide fluorophore 5-dimethylamino-1-naphthalene sulfonamide to human carbonic anhydrase II. Implications for the development of a zinc biosensor.
J.Biol.Chem., 271, 1996
|
|
3Q9C
 
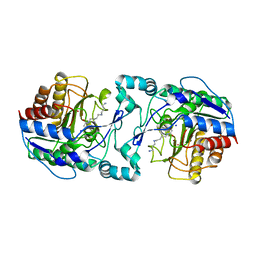 | |
1S8O
 
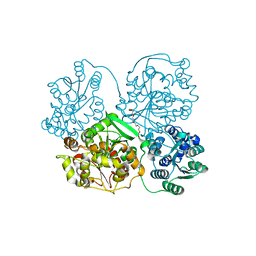 | | Human soluble Epoxide Hydrolase | | Descriptor: | HEXAETHYLENE GLYCOL, epoxide hydrolase 2, cytoplasmic | | Authors: | Gomez, G.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2004-02-03 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human epoxide hydrolase reveals mechanistic inferences on bifunctional catalysis in epoxide and phosphate ester hydrolysis
Biochemistry, 43, 2004
|
|
1YDD
 
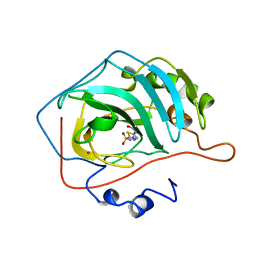 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
1YDA
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
1P42
 
 | | Crystal structure of Aquifex aeolicus LpxC Deacetylase (Zinc-Inhibited Form) | | Descriptor: | MYRISTIC ACID, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Whittington, D.A, Rusche, K.M, Shin, H, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2003-04-21 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of LpxC, a Zinc-Dependent Deacetylase Essential for Endotoxin Biosynthesis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1YDB
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
1RJ5
 
 | | Crystal Structure of the Extracellular Domain of Murine Carbonic Anhydrase XIV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Whittington, D.A, Grubb, J.H, Waheed, A, Shah, G.N, Sly, W.S, Christianson, D.W. | | Deposit date: | 2003-11-18 | | Release date: | 2004-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Expression, assay, and structure of the extracellular domain of murine carbonic anhydrase XIV: implications for selective inhibition of membrane-associated isozymes.
J.Biol.Chem., 279, 2004
|
|
1OKN
 
 | | CARBONIC ANHYDRASE II COMPLEX WITH THE 1OKN INHIBITOR 4-SULFONAMIDE-[1-(4-N-(5-FLUORESCEIN THIOUREA)BUTANE)] | | Descriptor: | 4-SULFONAMIDE-[4-(THIOMETHYLAMINOBUTANE)]BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Elbaum, D, Nair, S.K, Patchan, M.W, Thompson, R.B, Christianson, D.W. | | Deposit date: | 1996-06-25 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of a sulfonamide probe for fluorescence anisotropy detection of zinc with a carbonic anhydrase-based biosensor.
J.Am.Chem.Soc., 118, 1996
|
|
1R1O
 
 | |
1AS4
 
 | | CLEAVED ANTICHYMOTRYPSIN A349R | | Descriptor: | ACETATE ION, ANTICHYMOTRYPSIN | | Authors: | Lukacs, C.M, Christianson, D.W. | | Deposit date: | 1997-08-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering an anion-binding cavity in antichymotrypsin modulates the "spring-loaded" serpin-protease interaction.
Biochemistry, 37, 1998
|
|
1YDC
 
 | |
1EK1
 
 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE COMPLEXED WITH CIU INHIBITOR | | Descriptor: | EPOXIDE HYDROLASE, N-CYCLOHEXYL-N'-(4-IODOPHENYL)UREA | | Authors: | Argiriadi, M.A, Morisseau, C, Goodrow, M.H, Dowdy, D.L, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2000-03-06 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Binding of alkylurea inhibitors to epoxide hydrolase implicates active site tyrosines in substrate activation.
J.Biol.Chem., 275, 2000
|
|
1EK2
 
 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE COMPLEXED WITH CDU INHIBITOR | | Descriptor: | EPOXIDE HYDROLASE, N-CYCLOHEXYL-N'-DECYLUREA | | Authors: | Argiriadi, M.A, Morisseau, C, Goodrow, M.H, Dowdy, D.L, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2000-03-06 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Binding of alkylurea inhibitors to epoxide hydrolase implicates active site tyrosines in substrate activation.
J.Biol.Chem., 275, 2000
|
|
1RJ6
 
 | | Crystal Structure of the Extracellular Domain of Murine Carbonic Anhydrase XIV in Complex with Acetazolamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase XIV, ... | | Authors: | Whittington, D.A, Grubb, J.H, Waheed, A, Shah, G.N, Sly, W.S, Christianson, D.W. | | Deposit date: | 2003-11-18 | | Release date: | 2004-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Expression, assay, and structure of the extracellular domain of murine carbonic anhydrase XIV: implications for selective inhibition of membrane-associated isozymes.
J.Biol.Chem., 279, 2004
|
|
1OKM
 
 | | CARBONIC ANHYDRASE II COMPLEX WITH THE 1OKM INHIBITOR 4-SULFONAMIDE-[1-(4-AMINOBUTANE)]BENZAMIDE | | Descriptor: | 4-SULFONAMIDE-[1-(4-AMINOBUTANE)]BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Elbaum, D, Nair, S.K, Patchan, M.W, Thompson, R.B, Christianson, D.W. | | Deposit date: | 1996-06-25 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design of a sulfonamide probe for fluorescence anisotropy detection of zinc with a carbonic anhydrase-based biosensor.
J.Am.Chem.Soc., 118, 1996
|
|
3SL0
 
 | | Crystal Structure of P. falciparum arginase complexed with 2-amino-6-borono-2-(difluoromethyl)hexanoic acid | | Descriptor: | 2-(difluoromethyl)-6-(dihydroxyboranyl)-L-norleucine, Arginase, MANGANESE (II) ION | | Authors: | Dowling, D.P, Ilies, M, Christianson, D.W. | | Deposit date: | 2011-06-23 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Binding of alpha , alpha-disubstituted amino acids to arginase suggests new avenues for inhibitor design.
J.Med.Chem., 54, 2011
|
|
3RLA
 
 | |
3SL1
 
 | | Crystal Structure of P. falciparum arginase complexed with 2-amino-6-borono-2-methylhexanoic acid | | Descriptor: | 6-(dihydroxyboranyl)-2-methyl-L-norleucine, Arginase, MANGANESE (II) ION | | Authors: | Dowling, D.P, Ilies, M, Christianson, D.W. | | Deposit date: | 2011-06-23 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Binding of alpha , alpha-disubstituted amino acids to arginase suggests new avenues for inhibitor design.
J.Med.Chem., 54, 2011
|
|
3TH7
 
 | |
3TF3
 
 | |
