7C4Z
 
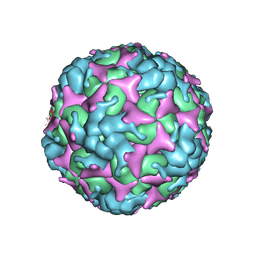 | | Cryo-EM structure of empty Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C4T
 
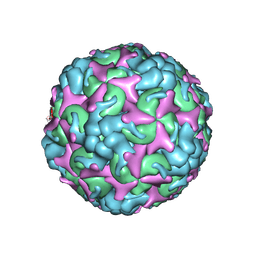 | | Cryo-EM structure of A particle Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C4W
 
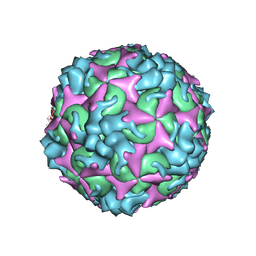 | | Cryo-EM structure of A particle Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7CP2
 
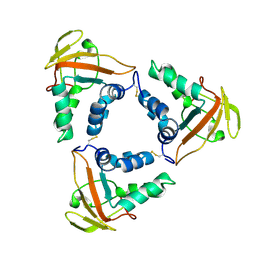 | | Crystal structure of the African swine fever virus core shell protein p15 | | Descriptor: | CP530R | | Authors: | Liu, K.F, Meng, Y.M, Chai, Y, Li, L.J, Sun, H, Gao, G.F, Tan, S.G, Qi, J.X. | | Deposit date: | 2020-08-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of the African swine fever virus core shell protein p15
Biosaf Health, 2021
|
|
7CU5
 
 | | N-Glycosylation of PD-1 and glycosylation dependent binding of PD-1 specific monoclonal antibody camrelizumab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Programmed cell death protein 1, ... | | Authors: | Liu, K.F, Tan, S.G, Jin, W.J, Guan, J.W, Wang, W.L, Sun, H, Qi, J.X, Yan, J.H, Chai, Y, Wang, Z.F, Chu, X.D, Gao, G.F. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | N-glycosylation of PD-1 promotes binding of camrelizumab.
Embo Rep., 21, 2020
|
|
7DEA
 
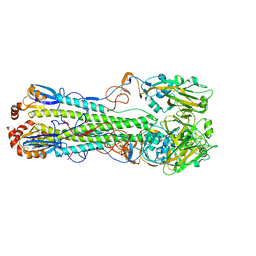 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck Northern China/22/2017 (H5N6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Sun, H, Sun, H, Song, J, Zhang, W, Wei, X, Qi, J, Gao, G.F, Liu, J. | | Deposit date: | 2020-11-03 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
7DEB
 
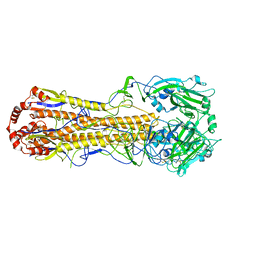 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck/Eastern China/L0230/2010 (H5N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Sun, H, Sun, H, Song, J, Zhang, W, Qi, J, Gao, G.F, Liu, J. | | Deposit date: | 2020-11-03 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
7CIR
 
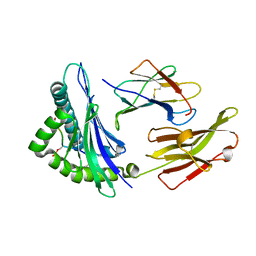 | | Peptide phosphorylation modification of MHC class I molecules | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SER-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CIQ
 
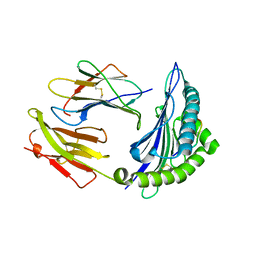 | | Phosphorylation modification of MHC I polypeptide | | Descriptor: | ARG-ARG-PHE-SER-ARG-SER-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CIS
 
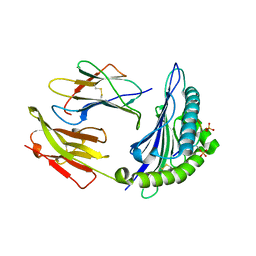 | | Peptide modification of MHC class I molecules | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SEP-PRO-ILE-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7BZT
 
 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C02
 
 | | Crystal structure of dimeric MERS-CoV receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Dai, L, Qi, J, Gao, G.F. | | Deposit date: | 2020-04-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS.
Cell, 182, 2020
|
|
7BZN
 
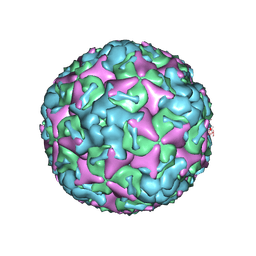 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7CZF
 
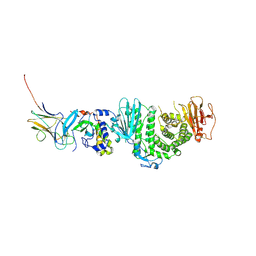 | | Crystal structure of Kaposi Sarcoma associated herpesvirus (KSHV ) gHgL in complex with the ligand binding domian (LBD) of EphA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Su, C, Wu, L.L, Song, H, Chai, Y, Qi, J.X, Yan, J.H, Gao, G.F. | | Deposit date: | 2020-09-08 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis of EphA2 recognition by gHgL from gammaherpesviruses.
Nat Commun, 11, 2020
|
|
7DDP
 
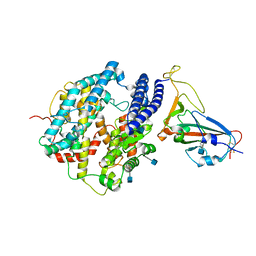 | | Cryo-EM structure of human ACE2 and GX/P2V/2017 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Niu, S, Wang, J, Wang, H.W, Qi, J.X, Wang, Q.H, Gao, G.F. | | Deposit date: | 2020-10-29 | | Release date: | 2021-05-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin.
Embo J., 40, 2021
|
|
7DDO
 
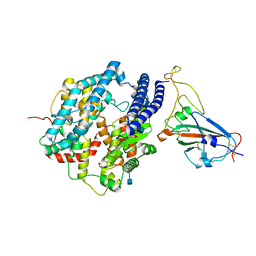 | | Cryo-EM structure of human ACE2 and GD/1/2019 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Niu, S, Wang, J, Wang, H.W, Qi, J.X, Wang, Q.H, Gao, G.F. | | Deposit date: | 2020-10-29 | | Release date: | 2021-05-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin.
Embo J., 40, 2021
|
|
7E57
 
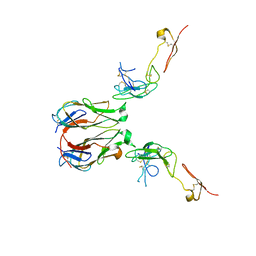 | | Crystal structure of murine GITR-GITRL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, Tumor necrosis factor receptor superfamily member 18, ... | | Authors: | Zhao, M, Tan, S, Fu, L, Chai, Y, Qi, J, Gao, G.F. | | Deposit date: | 2021-02-18 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Atypical TNF-TNFR superfamily binding interface in the GITR-GITRL complex for T cell activation.
Cell Rep, 36, 2021
|
|
7EKE
 
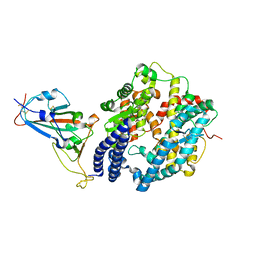 | | Structure of SARS-CoV-2 spike receptor-binding domain F486L mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKG
 
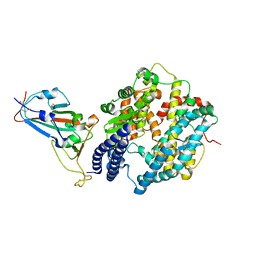 | | Structure of SARS-CoV-2 Beta variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKH
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKF
 
 | | Structure of SARS-CoV-2 Alpha variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKC
 
 | | Structure of SARS-CoV-2 Gamma variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7BZU
 
 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7E5N
 
 | | crystal structure of cis assembled TROP-2 | | Descriptor: | Tumor-associated calcium signal transducer 2 | | Authors: | Sun, M, Zhang, H, Chai, Y, Qi, J, Gao, G.F, Tan, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the cis and trans assembly of human trophoblast cell surface antigen 2.
Iscience, 24, 2021
|
|
7E5M
 
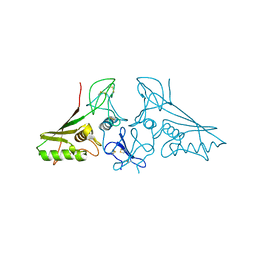 | | crystal structure of trans assembled human TROP-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor-associated calcium signal transducer 2 | | Authors: | Sun, M, Zhang, H, Chai, Y, Qi, J, Gao, G.F, Tan, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the cis and trans assembly of human trophoblast cell surface antigen 2.
Iscience, 24, 2021
|
|
