7YI8
 
 | | Cryo-EM structure of SAH-bound MTA1-MTA9-p1-p2 complex | | Descriptor: | MT-a70 family protein, MTA9, P1, ... | | Authors: | Yan, J.J, Guan, Z.Y, Liu, F.Q, Yan, X.H, Hou, M.J, Yin, P. | | Deposit date: | 2022-07-15 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into DNA N 6 -adenine methylation by the MTA1 complex.
Cell Discov, 9, 2023
|
|
7YI9
 
 | | Cryo-EM structure of SAM-bound MTA1-MTA9-p1-p2 complex | | Descriptor: | MT-a70 family protein, MTA9, P1, ... | | Authors: | Yan, J.J, Guan, Z.Y, Liu, F.Q, Yan, X.H, Hou, M.J, Yin, P. | | Deposit date: | 2022-07-15 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into DNA N 6 -adenine methylation by the MTA1 complex.
Cell Discov, 9, 2023
|
|
7YGL
 
 | |
7YHL
 
 | |
7YGH
 
 | |
3VOD
 
 | | Crystal Structure of mutant MarR C80S from E.coli | | Descriptor: | Multiple antibiotic resistance protein marR | | Authors: | Lou, H, Zhu, R, Hao, Z. | | Deposit date: | 2012-01-21 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The multiple antibiotic resistance regulator MarR is a copper sensor in Escherichia coli.
Nat.Chem.Biol., 10, 2014
|
|
3WD5
 
 | | Crystal structure of TNFalpha in complex with Adalimumab Fab fragment | | Descriptor: | Adalimumab Heavy Chain, Adalimumab Light Chain, Tumor necrosis factor | | Authors: | Hu, S, Liang, S.Y, Guo, Y.J, Lou, Z.Y. | | Deposit date: | 2013-06-06 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Comparison of the inhibition mechanisms of adalimumab and infliximab in treating tumor necrosis factor alpha-associated diseases from a molecular view
J.Biol.Chem., 288, 2013
|
|
8VQQ
 
 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule 6S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-01-19 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VQM
 
 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule 6R prime | | Descriptor: | (S~1~S,3R)-N-{3,5-dichloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-3-(dimethylamino)pyrrolidine-1-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-01-18 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VQL
 
 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule 6S prime | | Descriptor: | (S~1~S,3S)-N-{3,5-dichloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-3-(dimethylamino)pyrrolidine-1-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-01-18 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VQN
 
 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule 6R | | Descriptor: | (S~1~S,3R)-N-{3-chloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-3-(dimethylamino)pyrrolidine-1-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-01-18 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7E44
 
 | | Crystal structure of NudC complexed with dpCoA | | Descriptor: | DEPHOSPHO COENZYME A, NADH pyrophosphatase, ZINC ION | | Authors: | Zhou, W, Guan, Z.Y, Yin, P, Zhang, D.L. | | Deposit date: | 2021-02-10 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into dpCoA-RNA decapping by NudC.
Rna Biol., 18, 2021
|
|
7F6L
 
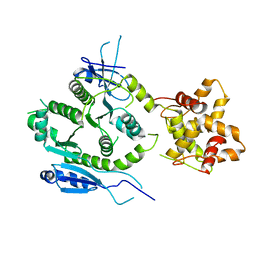 | | Crystal structure of human MUS81-EME2 complex | | Descriptor: | Crossover junction endonuclease MUS81, Probable crossover junction endonuclease EME2 | | Authors: | Hua, Z.K, Zhang, D.P, Yuan, C, Lin, Z.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human MUS81-EME2 complex.
Structure, 30, 2022
|
|
8GKF
 
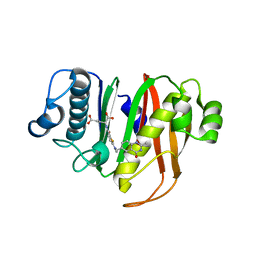 | |
8H3U
 
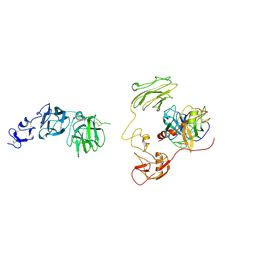 | | Inhibitor-bound EP, polyA model | | Descriptor: | Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain | | Authors: | Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-10-09 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
8H3S
 
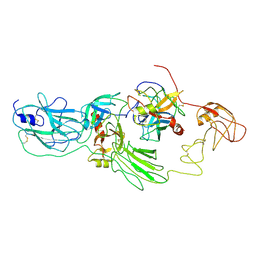 | | Substrate-bound EP, polyA model | | Descriptor: | Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain, Serine protease 1 | | Authors: | Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-10-09 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
8HFY
 
 | | SARS-CoV-2 Omicron BA.1 spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HFX
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HG0
 
 | | Cryo-EM structure of SARS-CoV-2 prototype spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HFZ
 
 | | Cryo-EM structure of SARS-CoV-2 prototype spike protein in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HTW
 
 | |
8IM7
 
 | | Human gamma-secretase treated with ganglioside GM1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, R, Yang, G, Shi, Y. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Preferential Regulation of Gamma-Secretase-Mediated Cleavage of APP by Ganglioside GM1 Reveals a Potential Therapeutic Target for Alzheimer's Disease.
Adv Sci, 10, 2023
|
|
8HR2
 
 | | Ternary Crystal Complex Structure of RBD with NB1B5 and NB1C6 | | Descriptor: | NB1B5, NB1C6, Spike protein S1 | | Authors: | Sun, Z. | | Deposit date: | 2022-12-14 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure basis of two nanobodies neutralizing SARS-CoV-2 Omicron variant by targeting ultra-conservative epitopes.
J.Struct.Biol., 215, 2023
|
|
8IFZ
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2023-02-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8IFY
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 spike protein in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2023-02-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
