5XZI
 
 | |
5XZJ
 
 | |
9KKH
 
 | | High resolution structure of Ferredoxin-NADP+ reductase from maize root - Reduced form, low X-ray dose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Uenaka, M, Ohnishi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2024-11-13 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Redox-dependent hydrogen-bond network rearrangement of ferredoxin-NADP + reductase revealed by high-resolution X-ray and neutron crystallography.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9KK7
 
 | | Neutron structure of Ferredoxin-NADP+ reductase from maize root -Reduced form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Uenaka, M, Ohnishi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2024-11-13 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | Redox-dependent hydrogen-bond network rearrangement of ferredoxin-NADP + reductase revealed by high-resolution X-ray and neutron crystallography.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9KKC
 
 | | Neutron structure of Ferredoxin-NADP+ reductase from maize root -Oxidized form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Uenaka, M, Ohnishi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2024-11-13 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | Redox-dependent hydrogen-bond network rearrangement of ferredoxin-NADP + reductase revealed by high-resolution X-ray and neutron crystallography.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9KKG
 
 | | High resolution structure of Ferredoxin-NADP+ reductase from maize root - Oxidized form, low X-ray dose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Uenaka, M, Ohnishi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2024-11-13 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Redox-dependent hydrogen-bond network rearrangement of ferredoxin-NADP + reductase revealed by high-resolution X-ray and neutron crystallography.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
6AX6
 
 | | The crystal structure of a lysyl hydroxylase from Acanthamoeba polyphaga mimivirus | | Descriptor: | FE (II) ION, IODIDE ION, Procollagen lysyl hydroxylase and glycosyltransferase | | Authors: | Guo, H, Tsai, C, Miller, M.D, Alvarado, S, Tainer, J.A, Phillips Jr, G.N, Kurie, J.M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding.
Nat Commun, 9, 2018
|
|
6AX7
 
 | | The crystal structure of a lysyl hydroxylase from Acanthamoeba polyphaga mimivirus | | Descriptor: | FE (II) ION, Procollagen lysyl hydroxylase and glycosyltransferase | | Authors: | Guo, H, Tsai, C, Miller, M.D, Alvarado, S, Tainer, J.A, Phillips Jr, G.N, Kurie, J.M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding.
Nat Commun, 9, 2018
|
|
5H06
 
 | | Crystal structure of AmyP in complex with maltose | | Descriptor: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | He, C, Liu, Y. | | Deposit date: | 2016-10-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
3WGN
 
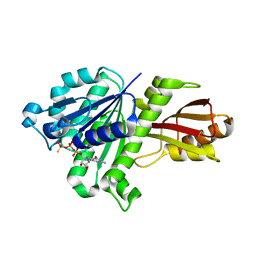 | | STAPHYLOCOCCUS AUREUS FTSZ bound with GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein FtsZ | | Authors: | Matsui, T, Mogi, N, Tanaka, I, Yao, M. | | Deposit date: | 2013-08-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Structural change in FtsZ Induced by intermolecular interactions between bound GTP and the T7 loop
J.Biol.Chem., 289, 2014
|
|
3WY2
 
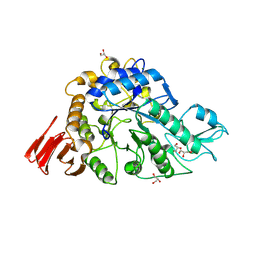 | | Crystal structure of alpha-glucosidase in complex with glucose | | Descriptor: | Alpha-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
3WY3
 
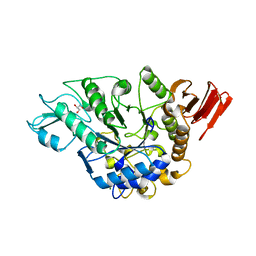 | | Crystal structure of alpha-glucosidase mutant D202N in complex with glucose and glycerol | | Descriptor: | Alpha-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
8XY4
 
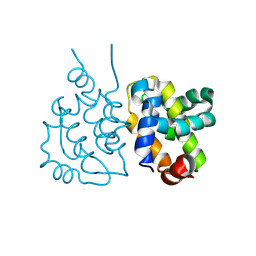 | | Crystal structure of VACV N1 protein | | Descriptor: | N1L | | Authors: | Ni, X.C, Lei, J. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into MPXV P1 protein and its orthologs reveal conformational dynamics and a conserved antiviral pocket.
Emerg Microbes Infect, 14, 2025
|
|
8XY1
 
 | | Crystal structure of MPXV P1 protein | | Descriptor: | Protein OPG035 | | Authors: | Ni, X.C, Lei, J. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.575 Å) | | Cite: | Structural insights into MPXV P1 protein and its orthologs reveal conformational dynamics and a conserved antiviral pocket.
Emerg Microbes Infect, 14, 2025
|
|
8XY3
 
 | | Crystal structure of VARV P1 protein | | Descriptor: | P1L | | Authors: | Ni, X.C, Lei, J. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into MPXV P1 protein and its orthologs reveal conformational dynamics and a conserved antiviral pocket.
Emerg Microbes Infect, 14, 2025
|
|
8XY2
 
 | | Crystal structure of MPXV P1 protein S87P mutant | | Descriptor: | Protein OPG035 | | Authors: | Ni, X.C, Lei, J. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural insights into MPXV P1 protein and its orthologs reveal conformational dynamics and a conserved antiviral pocket.
Emerg Microbes Infect, 14, 2025
|
|
3V06
 
 | | Crystal structure of S-6'-Me-3'-fluoro hexitol nucleic acid | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(F5H)P*AP*CP*GP*C)-3'), STRONTIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Insights from crystal structures into the opposite effects on RNA affinity caused by the s- and R-6'-methyl backbone modifications of 3'-fluoro hexitol nucleic Acid.
Biochemistry, 51, 2012
|
|
3V07
 
 | | Crystal structure of R-6'-Me-3'-fluoro hexitol nucleic acid | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(F6H)P*AP*CP*GP*C)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Insights from crystal structures into the opposite effects on RNA affinity caused by the s- and R-6'-methyl backbone modifications of 3'-fluoro hexitol nucleic Acid.
Biochemistry, 51, 2012
|
|
8FLP
 
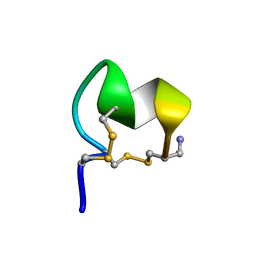 | | NMR Solution Structure of LvIC analogue | | Descriptor: | Alpha-conotoxin LvIC analogue | | Authors: | Harvey, P.J, Craik, D.J. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Discovery, Characterization, and Engineering of LvIC, an alpha 4/4-Conotoxin That Selectively Blocks Rat alpha 6/ alpha 3 beta 4 Nicotinic Acetylcholine Receptors.
J.Med.Chem., 66, 2023
|
|
8JT8
 
 | | Crystal structure of 5-HT2AR in complex with (R)-IHCH-7179 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Chen, Z, Fan, L, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
8JT6
 
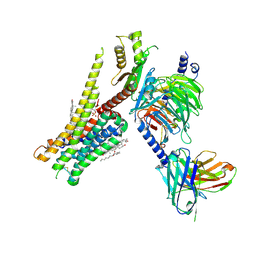 | | 5-HT1A-Gi in complex with compound (R)-IHCH-7179 | | Descriptor: | 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, Z, Xu, P, Huang, S, Xu, H.E, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
3R0H
 
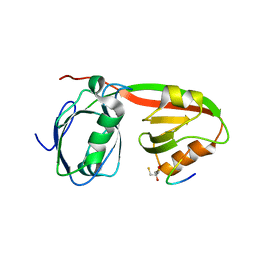 | | Structure of INAD PDZ45 in complex with NG2 peptide | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Inactivation-no-after-potential D protein, ... | | Authors: | Wei, Z, Liu, W, Zhang, M. | | Deposit date: | 2011-03-08 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The INAD scaffold is a dynamic, redox-regulated modulator of signaling in the Drosophila eye
Cell(Cambridge,Mass.), 145, 2011
|
|
3WY1
 
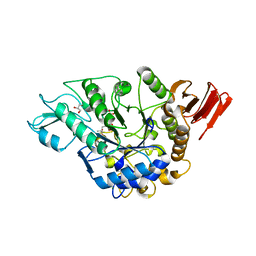 | | Crystal structure of alpha-glucosidase | | Descriptor: | (3R,5R,7R)-octane-1,3,5,7-tetracarboxylic acid, Alpha-glucosidase, GLYCEROL, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
3WY4
 
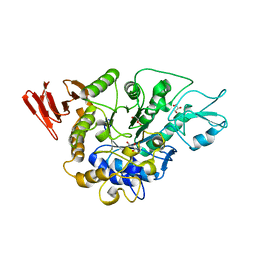 | | Crystal structure of alpha-glucosidase mutant E271Q in complex with maltose | | Descriptor: | Alpha-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
7M8K
 
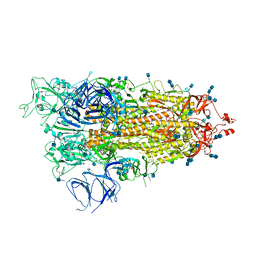 | | Cryo-EM structure of Brazil (P.1) SARS-CoV-2 spike glycoprotein variant in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L, Ho, D.D. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization.
Cell Host Microbe, 29, 2021
|
|
