4ZMI
 
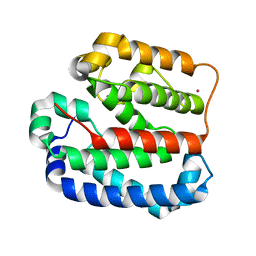 | | Crystal structure of the Helical domain of S. pombe Taz1 | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Telomere length regulator taz1 | | Authors: | Deng, W, Wu, J, Wang, F, Lei, M. | | Deposit date: | 2015-05-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fission yeast telomere-binding protein Taz1 is a functional but not a structural counterpart of human TRF1 and TRF2.
Cell Res., 25, 2015
|
|
6DYA
 
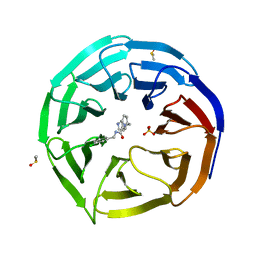 | | WDR5 in complex with a WIN site inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3,5-dichlorophenyl)methyl]-3-[(1H-imidazol-1-yl)methyl]benzamide, SULFATE ION, ... | | Authors: | Phan, J, Wang, F, Fesik, S.W. | | Deposit date: | 2018-07-01 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Displacement of WDR5 from Chromatin by a WIN Site Inhibitor with Picomolar Affinity.
Cell Rep, 26, 2019
|
|
4ZMK
 
 | |
4ZWV
 
 | | Crystal Structure of Aminotransferase AtmS13 from Actinomadura melliaura | | Descriptor: | GLYCEROL, Putative aminotransferase | | Authors: | Kim, Y, Bigelow, L, Endres, M, Wang, F, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-19 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
5W5F
 
 | | Cryo-EM structure of the T4 tail tube | | Descriptor: | Tail tube protein gp19 | | Authors: | Zheng, W, Wang, F, Taylor, N.M, Guerrero-Ferreira, R.C, Leiman, P.G, Egelman, E.H. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Refined Cryo-EM Structure of the T4 Tail Tube: Exploring the Lowest Dose Limit.
Structure, 25, 2017
|
|
3RLR
 
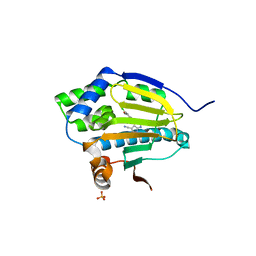 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-2,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-2,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RLQ
 
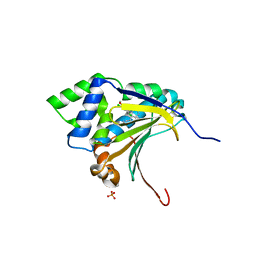 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-2-methyl-7H-pyrrolo[2,3-d]pyrimidine-5- carbonitrile | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-2-methyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RLP
 
 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6BNS
 
 | | STRUCTURE OF HUMAN PREGNANE X RECEPTOR LIGAND BINDING DOMAIN BOUND TETHERED WITH SRC co-activator peptide and Compound 25a AKA BICYCLIC HEXAFLUOROISOPROPYL 2 ALCOHOL SULFONAMIDES | | Descriptor: | 2-[(2S)-4-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)-3,4-dihydro-2H-1,4-benzothiazin-2-yl]-N-(2-hydroxy-2-methylpropyl)acetamide, Nuclear receptor subfamily 1 group I member 2,Nuclear receptor coactivator 1 Chimera | | Authors: | DHAR, T.G, GONG, H, WEINSTEIN, D.S, LU, Z, DUAN, J.J.W, STACHURA, S, HAQUE, L, KARMAKAR, A, HEMAGIRI, H, RAUT, D.K, GUPTA, A.K, KHAN, J.A, SACK, J.S, CAMAC, D.M, PUDZIANOWSKI, A.A, WU, D.R, YARDE, M, SHEN, D.R, BOROWSKI, V, XIE, J.H, SUN, H, ARIENZO, C.D, DABROS, M, GALELLA, M.A, WANG, F, WEIGELT, C.A, ZHAO, Q, FOSTER, W, SOMERVILLE, J.E, SALTER-CID, L.M, BARRISH, J.C, CARTER, P.H. | | Deposit date: | 2017-11-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Identification of bicyclic hexafluoroisopropyl alcohol sulfonamides as retinoic acid receptor-related orphan receptor gamma (ROR gamma /RORc) inverse agonists. Employing structure-based drug design to improve pregnane X receptor (PXR) selectivity.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6C53
 
 | | Cryo-EM structure of the Type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Zheng, W, Wang, F, Luna-Rico, A, Francetic, O, Hultgren, S.J, Egelman, E.H. | | Deposit date: | 2018-01-13 | | Release date: | 2018-01-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Functional role of the type 1 pilus rod structure in mediating host-pathogen interactions.
Elife, 7, 2018
|
|
8TIB
 
 | | Cryo-EM of tri-pilus from S. islandicus REY15A | | Descriptor: | DUF973 family protein | | Authors: | Eastep, G.N, Liu, J, Rich-New, S.T, Egelman, E.H, Krupovic, M, Wang, F. | | Deposit date: | 2023-07-19 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Cryo-EM of tri-pilus from S. islandicus REY15A
To Be Published
|
|
8TIF
 
 | | Cryo-EM of mono-pilus from S. islandicus REY15A | | Descriptor: | DUF973 family protein | | Authors: | Eastep, G.N, Liu, J, Rich-New, S.T, Egelman, E.H, Krupovic, M, Wang, F. | | Deposit date: | 2023-07-19 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Cryo-EM of mono-pilus from S. islandicus REY15A
To Be Published
|
|
3TLQ
 
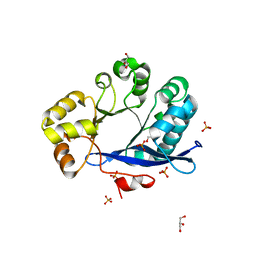 | | Crystal structure of EAL-like domain protein YdiV | | Descriptor: | GLYCEROL, PHOSPHATE ION, Regulatory protein YdiV | | Authors: | Li, B, Li, N, Wang, F, Guo, L, Liu, C, Zhu, D, Xu, S, Gu, L. | | Deposit date: | 2011-08-30 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility
Nucleic Acids Res., 40, 2012
|
|
6HQE
 
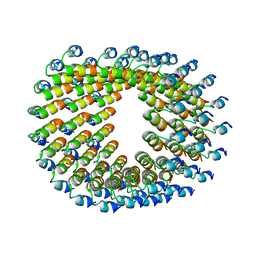 | | Cryo-EM of self-assembly peptide filament LRV_M3delta1 | | Descriptor: | peptide LRV_M3delta1 | | Authors: | Osinski, T, Wang, F, Hughes, S.A, Kreutzberger, M.A.B, Conticello, V.P, Egelman, E.H. | | Deposit date: | 2018-09-24 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Ambidextrous helical nanotubes from self-assembly of designed helical hairpin motifs.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K6L
 
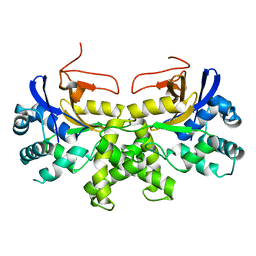 | | YGL082W-catalytic domain | | Descriptor: | pseudo deubiquitinase | | Authors: | Lu, L.N, Wang, F. | | Deposit date: | 2019-06-03 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Inactivity of YGL082W in vitro due to impairment of conformational change in the catalytic center loop
Sci China Chem, 2019
|
|
5F0U
 
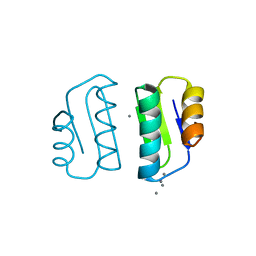 | | Crystal structure of Gold binding protein | | Descriptor: | Putative copper chaperone, SILVER ION | | Authors: | Wei, W, Wang, F, Zhao, J. | | Deposit date: | 2015-11-28 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of tetrasilver bound to GolB at 1.70 Angstroms resolution
To Be Published
|
|
4MRM
 
 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist phaclofen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-17 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4LI5
 
 | | EGFR-K IN COMPLEX WITH N-[3-[[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]-4-methoxy-phenyl] Prop-2-enamide | | Descriptor: | Epidermal growth factor receptor, N-(3-{[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino}-4-methoxyphenyl)propanamide, SODIUM ION | | Authors: | Debreczeni, J.E, Seiffert, G.B, Kiefersauer, R, Augustin, M, Nagel, S, Ward, R, Anderton, M, Ashton, S, Bethel, P, Box, M, Butterworth, S, Colclough, N, Chroley, C, Chuaqui, C, Cross, D, Eberlein, C, Finlay, R, Hill, G, Grist, M, Klinowska, T, Lane, C, Martin, S, Orme, J, Smith, P, Wang, F, Waring, M. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure- and Reactivity-Based Development of Covalent Inhibitors of the Activating and Gatekeeper Mutant Forms of the Epidermal Growth Factor Receptor (EGFR).
J.Med.Chem., 56, 2013
|
|
5GZ2
 
 | | luciferase complex with 7-cy-L | | Descriptor: | (4S)-2-[6-(azepan-1-yl)-1,3-benzothiazol-2-yl]-4,5-dihydro-1,3-thiazole-4-carboxylic acid, Luciferin 4-monooxygenase | | Authors: | Su, J, Wang, F, Gu, L. | | Deposit date: | 2016-09-26 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | luciferase complex with 7-cy-L
To Be Published
|
|
4IR7
 
 | | Crystal Structure of Mtb FadD10 in Complex with Dodecanoyl-AMP | | Descriptor: | 5'-O-[(S)-(dodecanoyloxy)(hydroxy)phosphoryl]adenosine, Long chain fatty acid CoA ligase FadD10, MAGNESIUM ION | | Authors: | Liu, Z, Wang, F, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Mycobacterium tuberculosis FadD10 protein reveal a new type of adenylate-forming enzyme.
J.Biol.Chem., 288, 2013
|
|
4ISB
 
 | |
4M60
 
 | | Crystal structure of macrolide glycosyltransferases OleD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Oleandomycin glycosyltransferase, SODIUM ION | | Authors: | Olmos Jr, J.L, Martinez III, E, Wang, F, Helmich, K.E, Singh, S, Xu, W, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-04 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4MR8
 
 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist CGP35348 | | Descriptor: | (S)-(3-aminopropyl)(diethoxymethyl)phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-17 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4MS3
 
 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the endogenous agonist GABA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4MR7
 
 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist CGP54626 | | Descriptor: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-17 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
