5OR2
 
 | | Crystal structures of PYR1/HAB1 in complex with synthetic analogues of Abscisic Acid | | Descriptor: | (2~{Z},4~{E})-3-cyclopropyl-5-[(1~{S})-2,6,6-trimethyl-1-oxidanyl-4-oxidanylidene-cyclohex-2-en-1-yl]penta-2,4-dienoic acid, Abscisic acid receptor PYR1, MANGANESE (II) ION, ... | | Authors: | Freigang, J. | | Deposit date: | 2017-08-15 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the in Vitro and in Vivo SAR of Abscisic Acid - Exploring Unprecedented Variations of the Side Chain via Cross-Coupling-Mediated Syntheses
Eur.J.Org.Chem., 2018
|
|
1QUR
 
 | |
6I8G
 
 | |
5OR6
 
 | | Crystal structures of PYR1/HAB1 in complex with synthetic analogues of Abscisic Acid | | Descriptor: | (~{E})-3-(trifluoromethyl)-5-[(1~{S})-2,6,6-trimethyl-1-oxidanyl-4-oxidanylidene-cyclohex-2-en-1-yl]pent-2-en-4-ynoic acid, Abscisic acid receptor PYR1, MANGANESE (II) ION, ... | | Authors: | Freigang, J. | | Deposit date: | 2017-08-15 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the in Vitro and in Vivo SAR of Abscisic Acid - Exploring Unprecedented Variations of the Side Chain via Cross-Coupling-Mediated Syntheses
Eur.J.Org.Chem., 2018
|
|
2WBS
 
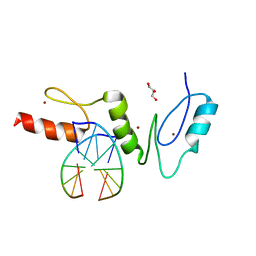 | | Crystal structure of the zinc finger domain of Klf4 bound to its target DNA | | Descriptor: | 5'-D(*GP*AP*GP*GP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*CP*CP*TP*CP)-3', GLYCEROL, ... | | Authors: | Zocher, G, Schuetz, A, Carstanjen, D, Heinemann, U. | | Deposit date: | 2009-03-03 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Klf4 DNA-Binding Domain Links to Self-Renewal and Macrophage Differentiation.
Cell.Mol.Life Sci., 68, 2011
|
|
3ZEP
 
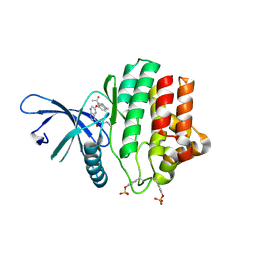 | | Crystal Structure of JAK3 Kinase Domain in Complex with a Pyrrolopyrazine-2-phenyl Ether Inhibitor | | Descriptor: | 2-[[(3R)-3-acetamido-2,3-dihydro-1H-inden-5-yl]oxy]-N-[(1S)-1-cyclopropylethyl]-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, GLYCEROL, TYROSINE-PROTEIN KINASE JAK3 | | Authors: | Kuglstatter, A, Jestel, A, Nagel, S, Boettcher, J, Blaesse, M. | | Deposit date: | 2012-12-06 | | Release date: | 2013-12-11 | | Last modified: | 2016-09-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of a Series of Novel 5H-Pyrrolo[2,3-B]Pyrazine-2-Phenyl Ethers, as Potent Jak3 Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2WBU
 
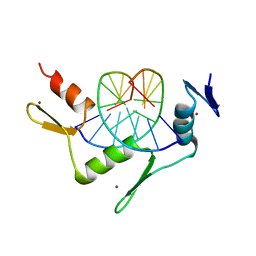 | | CRYSTAL STRUCTURE OF THE ZINC FINGER DOMAIN OF KLF4 BOUND TO ITS TARGET DNA | | Descriptor: | 5'-D(*DGP*DAP*DGP*DGP*DCP*DGP*DTP* DGP*DGP*DC)-3', 5'-D(*DGP*DCP*DCP*DAP*DCP*DGP*DCP* DCP*DTP*DC)-3', KRUEPPEL-LIKE FACTOR 4, ... | | Authors: | Schuetz, A, Zocher, G, Carstanjen, D, Heinemann, U. | | Deposit date: | 2009-03-05 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of the Klf4 DNA-Binding Domain Links to Self-Renewal and Macrophage Differentiation.
Cell.Mol.Life Sci., 68, 2011
|
|
2OB1
 
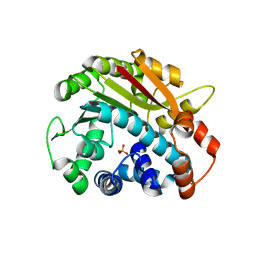 | | ppm1 with 1,8-ANS | | Descriptor: | Leucine carboxyl methyltransferase 1, PHOSPHATE ION | | Authors: | Groves, M.R. | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A method for the general identification of protein crystals in crystallization experiments using a noncovalent fluorescent dye.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6I8H
 
 | |
1TRQ
 
 | |
1TRP
 
 | |
3DNG
 
 | | Crystal structure of the complex between MMP-8 and a non-zinc chelating inhibitor | | Descriptor: | (5S)-5-(2-amino-2-oxoethyl)-4-oxo-N-[(3-oxo-3,4-dihydro-2H-1,4-benzoxazin-6-yl)methyl]-3,4,5,6,7,8-hexahydro[1]benzothieno[2,3-d]pyrimidine-2-carboxamide, CALCIUM ION, Neutrophil collagenase, ... | | Authors: | Pochetti, G, Montanari, R, Mazza, F. | | Deposit date: | 2008-07-02 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extra Binding Region Induced by Non-Zinc Chelating Inhibitors into the S(1)' Subsite of Matrix Metalloproteinase 8 (MMP-8)
J.Med.Chem., 52, 2009
|
|
3DPE
 
 | | Crystal structure of the complex between MMP-8 and a non-zinc chelating inhibitor | | Descriptor: | CALCIUM ION, N-{[2-(2-amino-3,4-dioxocyclobut-1-en-1-yl)-1,2,3,4-tetrahydroisoquinolin-7-yl]methyl}-4-oxo-3,5,6,8-tetrahydro-4H-thiopyrano[4',3':4,5]thieno[2,3-d]pyrimidine-2-carboxamide 7,7-dioxide, Neutrophil collagenase, ... | | Authors: | Pochetti, G, Montanari, R, Mazza, F. | | Deposit date: | 2008-07-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Extra Binding Region Induced by Non-Zinc Chelating Inhibitors into the S(1)' Subsite of Matrix Metalloproteinase 8 (MMP-8)
J.Med.Chem., 52, 2009
|
|
3DPF
 
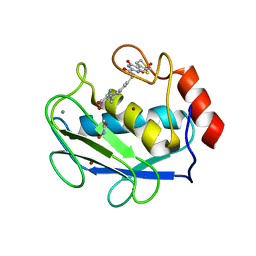 | | Crystal structure of the complex between MMP-8 and a non-zinc chelating inhibitor | | Descriptor: | ACETOHYDROXAMIC ACID, CALCIUM ION, N-{[2-(2-amino-3,4-dioxocyclobut-1-en-1-yl)-1,2,3,4-tetrahydroisoquinolin-7-yl]methyl}-4-oxo-3,5,6,8-tetrahydro-4H-thiopyrano[4',3':4,5]thieno[2,3-d]pyrimidine-2-carboxamide 7,7-dioxide, ... | | Authors: | Pochetti, G, Montanari, R, Mazza, F. | | Deposit date: | 2008-07-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Extra Binding Region Induced by Non-Zinc Chelating Inhibitors into the S(1)' Subsite of Matrix Metalloproteinase 8 (MMP-8)
J.Med.Chem., 52, 2009
|
|
4I6Q
 
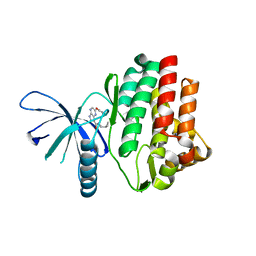 | | JAK3 kinase domain in complex with 2-Phenoxy-5H-pyrrolo[2,3-b]pyrazine-7-carboxylic acid ((S)-1-cyclopropyl-ethyl)-amide | | Descriptor: | 1-phenylurea, N-[(1S)-1-cyclopropylethyl]-2-phenoxy-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Kuglstatter, A, Shao, A. | | Deposit date: | 2012-11-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of a series of novel 5H-pyrrolo[2,3-b]pyrazine-2-phenyl ethers, as potent JAK3 kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6Q6Z
 
 | |
5DLV
 
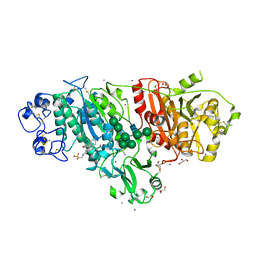 | | Crystal structure of Autotaxin (ENPP2) with tauroursodeoxycholic acid (TUDCA) | | Descriptor: | 2-{[(3alpha,5beta,7alpha,8alpha,14beta,17alpha)-3,7-dihydroxy-24-oxocholan-24-yl]amino}ethanesulfonic acid, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Keune, W.J, Heidebrecht, T, von Castelmur, E, Joosten, R.P, Perrakis, A. | | Deposit date: | 2015-09-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Steroid binding to Autotaxin links bile salts and lysophosphatidic acid signalling.
Nat Commun, 7, 2016
|
|
4N20
 
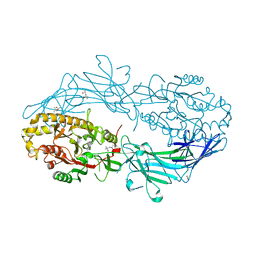 | | Crystal structure of Protein Arginine Deiminase 2 (0 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
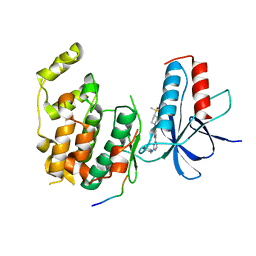
4IZY
 
 | |
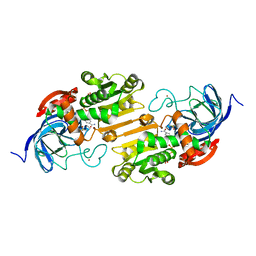
2OXI
 
 | |
3GOK
 
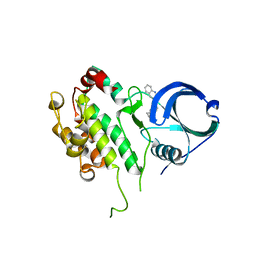 | | Binding site mapping of protein ligands | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP kinase-activated protein kinase 2 | | Authors: | Scheich, C. | | Deposit date: | 2009-03-19 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Binding site mapping of protein ligands
To be published
|
|
5MJ0
 
 | |
5MJ2
 
 | | Extracellular domain of human CD83 - rhombohedral crystal form after UV-RIP (S-SAD data) | | Descriptor: | CD83 antigen, DI(HYDROXYETHYL)ETHER | | Authors: | Klingl, S, Egerer-Sieber, C, Schmid, B, Weiler, S, Muller, Y.A. | | Deposit date: | 2016-11-29 | | Release date: | 2017-03-29 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of the Extracellular Domain of the Human Dendritic Cell Surface Marker CD83.
J. Mol. Biol., 429, 2017
|
|
5MIX
 
 | |
6HUM
 
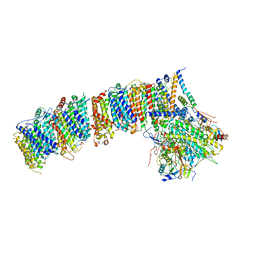 | | Structure of the photosynthetic complex I from Thermosynechococcus elongatus | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, IRON/SULFUR CLUSTER, ... | | Authors: | Schuller, J.M, Schuller, S.K, Kurisu, G, Engel, B.D, Nowaczyk, M.M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer.
Science, 363, 2019
|
|
