3IKA
 
 | |
4LX4
 
 | | Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein | | Authors: | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.556 Å) | | Cite: | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6JIF
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Pseudomonas sp. UW4 | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zheng, X, Guo, L, Li, D.F, Wu, B. | | Deposit date: | 2019-02-21 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of a highly active branched-chain amino acid aminotransferase from Pseudomonas sp. for efficient biosynthesis of chiral amino acids.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
7OTJ
 
 | | Crystal structure of Pif1 helicase from Candida albicans | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Rety, S, Xi, X.G. | | Deposit date: | 2021-06-10 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural study of the function of Candida Albicans Pif1.
Biochem.Biophys.Res.Commun., 567, 2021
|
|
4AEE
 
 | | CRYSTAL STRUCTURE OF MALTOGENIC AMYLASE FROM S.MARINUS | | Descriptor: | ALPHA AMYLASE, CATALYTIC REGION | | Authors: | Jung, T.Y, Park, C.H, Yoon, S.M, Park, S.H, Park, K.H, Woo, E.J. | | Deposit date: | 2012-01-10 | | Release date: | 2012-01-18 | | Last modified: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Association of Novel Domain in Active Site of Archaic Hyperthermophilic Maltogenic Amylase from Staphylothermus Marinus.
J.Biol.Chem., 287, 2012
|
|
3L0T
 
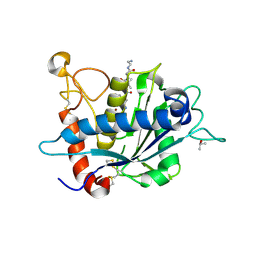 | | Crystal structure of catalytic domain of TACE with hydantoin inhibitor | | Descriptor: | Disintegrin and metalloproteinase domain-containing protein 17, ISOPROPYL ALCOHOL, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, ... | | Authors: | Orth, P. | | Deposit date: | 2009-12-10 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery and SAR of hydantoin TACE inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8I4Z
 
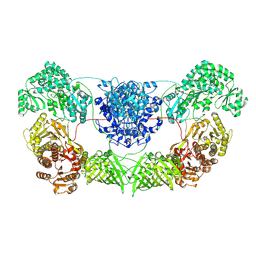 | | CalA3 with hydrolysis product | | Descriptor: | 11-oxidanylidene-11-(1~{H}-pyrrol-2-yl)undecanoic acid, Beta-ketoacyl-acyl-carrier-protein synthase I | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2023-01-21 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | C-N bond formation by a polyketide synthase.
Nat Commun, 14, 2023
|
|
8I4Y
 
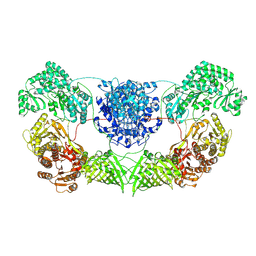 | | CalA3 complex structure with amidation product | | Descriptor: | 11-oxidanylidene-11-(1~{H}-pyrrol-2-yl)undecanoic acid, 3-HYDROXYANTHRANILIC ACID, Beta-ketoacyl-acyl-carrier-protein synthase I | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2023-01-21 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | C-N bond formation by a polyketide synthase.
Nat Commun, 14, 2023
|
|
7LYK
 
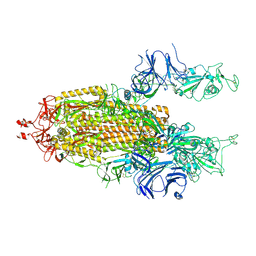 | |
7LYQ
 
 | |
7LYL
 
 | |
7LYO
 
 | |
7LYP
 
 | |
7LYN
 
 | |
7CBZ
 
 | | Crystal structure of T2R-TTL-A31 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[5-[4-[2-[4-(2-cyclopropylethanoyl)piperazin-1-yl]ethoxy]phenyl]pyridin-2-yl]-N-(phenylmethyl)ethanamide, CALCIUM ION, ... | | Authors: | Yang, J.H, Yan, W. | | Deposit date: | 2020-06-15 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Design, Synthesis, and Bioactivity Evaluation of Dual-Target Inhibitors of Tubulin and Src Kinase Guided by Crystal Structure.
J.Med.Chem., 64, 2021
|
|
3L0V
 
 | |
4FF5
 
 | |
8JEY
 
 | |
8IUM
 
 | | Cryo-EM structure of the tafluprost acid-bound human PTGFR-Gq complex | | Descriptor: | (~{Z})-7-[(1~{R},2~{R},3~{R},5~{S})-2-[(~{E})-3,3-bis(fluoranyl)-4-phenoxy-but-1-enyl]-3,5-bis(oxidanyl)cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, G subunit alpha (q), ... | | Authors: | Wu, C, Xu, Y, Xu, H.E. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Nat Commun, 14, 2023
|
|
8IUK
 
 | | Cryo-EM structure of the PGF2-alpha-bound human PTGFR-Gq complex | | Descriptor: | (Z)-7-[(1R,2R,3R,5S)-3,5-bis(oxidanyl)-2-[(E,3S)-3-oxidanyloct-1-enyl]cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, G subunit alpha (q), ... | | Authors: | Wu, C, Xu, Y, Xu, H.E. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Nat Commun, 14, 2023
|
|
8IUL
 
 | | Cryo-EM structure of the latanoprost-bound human PTGFR-Gq complex | | Descriptor: | Antibody fragment scFv16, G subunit alpha (q), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wu, C, Xu, Y, Xu, H.E. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Nat Commun, 14, 2023
|
|
8J5Z
 
 | | The cryo-EM structure of the TwOSC1 tetramer | | Descriptor: | Terpene cyclase/mutase family member, octyl beta-D-glucopyranoside | | Authors: | Ma, X, Yuru, T, Yunfeng, L, Jiang, T. | | Deposit date: | 2023-04-24 | | Release date: | 2023-11-01 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | Structural and Catalytic Insight into the Unique Pentacyclic Triterpene Synthase TwOSC.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8IXZ
 
 | |
8IY1
 
 | | Structure of Acb2 complexed with cAAG | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate, p26 | | Authors: | Cao, X.L, Xiao, Y, Feng, Y. | | Deposit date: | 2023-04-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phage anti-CBASS protein simultaneously sequesters cyclic trinucleotides and dinucleotides.
Mol.Cell, 84, 2024
|
|
8IY0
 
 | | Structure of Acb2 complexed with cAAA | | Descriptor: | [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate, p26 | | Authors: | Cao, X.L, Xiao, Y, Feng, Y. | | Deposit date: | 2023-04-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Phage anti-CBASS protein simultaneously sequesters cyclic trinucleotides and dinucleotides.
Mol.Cell, 84, 2024
|
|
