1DEA
 
 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
1ES9
 
 | | X-RAY CRYSTAL STRUCTURE OF R22K MUTANT OF THE MAMMALIAN BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASES (PAF-AH) | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE IB GAMMA SUBUNIT | | Authors: | McMullen, T.W.P, Li, J, Sheffield, P.J, Aoki, J, Martin, T.W, Arai, H, Inoue, K, Derewenda, Z.S. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The functional implications of the dimerization of the catalytic subunits of the mammalian brain platelet-activating factor acetylhydrolase (Ib).
Protein Eng., 13, 2000
|
|
1EIB
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT D313A COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-25 | | Release date: | 2001-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1EDQ
 
 | |
1EMR
 
 | | CRYSTAL STRUCTURE OF HUMAN LEUKEMIA INHIBITORY FACTOR (LIF) | | Descriptor: | LEUKEMIA INHIBITORY FACTOR | | Authors: | Robinson, R.C, Heath, J.K, Hawkins, N, Samal, B, Jones, E.Y, Betzel, C. | | Deposit date: | 2000-03-17 | | Release date: | 2001-03-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Species Variation in Receptor Binding Site Revealed by the Medium Resolution X-ray Structure of Human Leukemia Inhibitory Factor
to be published
|
|
1EHN
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT E315Q COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-22 | | Release date: | 2001-02-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1E5D
 
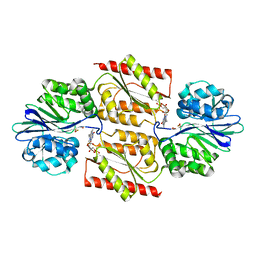 | | RUBREDOXIN OXYGEN:OXIDOREDUCTASE (ROO) FROM ANAEROBE DESULFOVIBRIO GIGAS | | Descriptor: | FLAVIN MONONUCLEOTIDE, MU-OXO-DIIRON, OXYGEN MOLECULE, ... | | Authors: | Frazao, C, Silva, G, Gomes, C.M, Matias, P, Coelho, R, Sieker, L, Macedo, S, Liu, M.Y, Oliveira, S, Teixeira, M, Xavier, A.V, Rodrigues-Pousada, C, Carrondo, M.A, Le Gall, J. | | Deposit date: | 2000-07-24 | | Release date: | 2000-11-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Dioxygen Reduction Enzyme from Desulfovibrio Gigas
Nat.Struct.Biol., 7, 2000
|
|
4LVC
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenosine | | Descriptor: | ACETATE ION, ADENOSINE, AMMONIUM ION, ... | | Authors: | Manszewski, T, Singh, K, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | An enzyme captured in two conformational states: crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3DT9
 
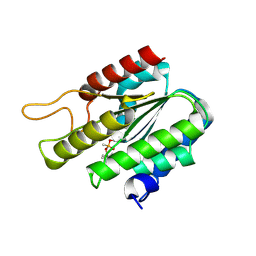 | | Crystal Structure of Bovin Brain Platelet Activating Factor Acetylhydrolase Covalently Inhibited by Soman | | Descriptor: | (1R)-1,2,2-TRIMETHYLPROPYL (R)-METHYLPHOSPHINATE, Brain Platelet-activating factor acetylhydrolase IB subunit alpha | | Authors: | Epstein, T.M, Samanta, U, Bahnson, B.J. | | Deposit date: | 2008-07-14 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of brain group-VIII phospholipase A2 in nonaged complexes with the organophosphorus nerve agents soman and sarin.
Biochemistry, 48, 2009
|
|
3DT8
 
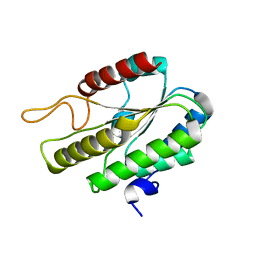 | |
3DT6
 
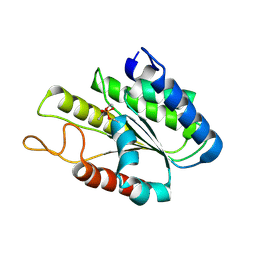 | |
5TTR
 
 | | LEU 55 PRO TRANSTHYRETIN CRYSTAL STRUCTURE | | Descriptor: | TRANSTHYRETIN | | Authors: | Sebastiao, M.P, Saraiva, M.J, Damas, A.M. | | Deposit date: | 1998-04-30 | | Release date: | 1999-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of amyloidogenic Leu55 --> Pro transthyretin variant reveals a possible pathway for transthyretin polymerization into amyloid fibrils.
J.Biol.Chem., 273, 1998
|
|
3AFP
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form I) | | Descriptor: | CADMIUM ION, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3A5U
 
 | | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex | | Descriptor: | DNA (31-MER), Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Manjunath, G.P, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2009-08-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex.
To be Published, 2009
|
|
5W19
 
 | | Tryptophan indole-lyase complex with oxindolyl-L-alanine | | Descriptor: | 1-carboxy-1-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]azaniumyl}-2-[(3R)-2-oxo-2,3-dihydro-1H-indol-3-yl]ethan-1-ide, POTASSIUM ION, Tryptophanase | | Authors: | Phillips, R.S, Wood, Z.A. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of Proteus vulgaris tryptophan indole-lyase complexed with oxindolyl-L-alanine: implications for the reaction mechanism.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4EKF
 
 | |
1FXW
 
 | |
3NUL
 
 | | Profilin I from Arabidopsis thaliana | | Descriptor: | GLYCEROL, PROFILIN I, SULFATE ION | | Authors: | Thorn, K, Christensen, H.E.M, Shigeta, R, Huddler, D, Chua, N.-H, Shalaby, L, Lindberg, U, Schutt, C.E. | | Deposit date: | 1996-11-27 | | Release date: | 1997-12-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a major allergen from plants.
Structure, 5, 1997
|
|
1UPD
 
 | | Oxidized STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 AT PH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Bento, I, Matias, P.M, Baptista, A.M, Da Costa, P.N, Van Dongen, W.M.A.M, Saraiva, L.M, Schneider, T.R, Soares, C.M, Carrondo, M.A. | | Deposit date: | 2003-09-29 | | Release date: | 2004-09-30 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Basis for Redox-Bohr and Cooperative Effects in Cytochrome C3 from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies of Oxidized and Reduced High-Resolution Structures at Ph 7.6
Proteins, 54, 2004
|
|
1X1V
 
 | | Structure Of Banana Lectin- Methyl-Alpha-Mannose Complex | | Descriptor: | HEXANE-1,6-DIOL, ZINC ION, lectin, ... | | Authors: | Singh, D.D, Saikrishnan, K, Kumar, P, Surolia, A, Sekar, K, Vijayan, M. | | Deposit date: | 2005-04-14 | | Release date: | 2005-11-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Unusual sugar specificity of banana lectin from Musa paradisiaca and its probable evolutionary origin. Crystallographic and modelling studies
Glycobiology, 15, 2005
|
|
1WAP
 
 | |
1UP9
 
 | | REDUCED STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 AT PH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C, SULFATE ION | | Authors: | Bento, I, Matias, P.M, Baptista, A.M, Da Costa, P.N, Van Dongen, W.M.A.M, Saraiva, L.M, Schneider, T.R, Soares, C.M, Carrondo, M.A. | | Deposit date: | 2003-09-29 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Molecular Basis for Redox-Bohr and Cooperative Effects in Cytochrome C3 from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies of Oxidized and Reduced High-Resolution Structures at Ph 7.6
Proteins, 54, 2004
|
|
1Z0W
 
 | | Crystal Structure of A. fulgidus Lon proteolytic domain at 1.2A resolution | | Descriptor: | CALCIUM ION, Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-02 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
1YCB
 
 | | DISTAL POCKET POLARITY IN LIGAND BINDING TO MYOGLOBIN: DEOXY AND CARBONMONOXY FORMS OF A THREONINE68 (E11) MUTANT INVESTIGATED BY X-RAY CRYSTALLOGRAPHY AND INFRARED SPECTROSCOPY | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cameron, A.D, Smerdon, S.J, Wilkinson, A.J, Habash, J, Helliwell, J.R. | | Deposit date: | 1993-08-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distal pocket polarity in ligand binding to myoglobin: deoxy and carbonmonoxy forms of a threonine68(E11) mutant investigated by X-ray crystallography and infrared spectroscopy.
Biochemistry, 32, 1993
|
|
1YCA
 
 | | DISTAL POCKET POLARITY IN LIGAND BINDING TO MYOGLOBIN: DEOXY AND CARBONMONOXY FORMS OF A THREONINE68 (E11) MUTANT INVESTIGATED BY X-RAY CRYSTALLOGRAPHY AND INFRARED SPECTROSCOPY | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cameron, A.D, Smerdon, S.J, Wilkinson, A.J, Habash, J, Helliwell, J.R. | | Deposit date: | 1993-08-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Distal pocket polarity in ligand binding to myoglobin: deoxy and carbonmonoxy forms of a threonine68(E11) mutant investigated by X-ray crystallography and infrared spectroscopy.
Biochemistry, 32, 1993
|
|
