6LHZ
 
 | | Structure of aerolysin-like protein (Bombina maxima) | | Descriptor: | aerolysin-like protein | | Authors: | Bian, X.L, Wang, Q.Q, Li, X, Teng, M.Q, Zhang, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A cellular endolysosome-modulating pore-forming protein from a toad is negatively regulated by its paralog under oxidizing conditions.
J.Biol.Chem., 295, 2020
|
|
5XSG
 
 | | Ultrahigh resolution structure of FUS (37-42) SYSGYS determined by MicroED | | Descriptor: | RNA-binding protein FUS | | Authors: | Luo, F, Gui, X, Zhou, H, Li, D, Li, X, Liu, C. | | Deposit date: | 2017-06-14 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.73 Å) | | Cite: | Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3QXL
 
 | | Crystal structure of the CDC25 Domain from Ral-specific Guanine-nucleotide Exchange Factor RalGPS1a | | Descriptor: | Ras-specific guanine nucleotide-releasing factor RalGPS1 | | Authors: | Peng, W, Xu, J, Guan, X, Sun, Y, Li, X, Zhang, X.C, Rao, Z. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.237 Å) | | Cite: | Structural study of the Cdc25 domain from Ral-specific guanine-nucleotide exchange factor RalGPS1a.
Protein Cell, 2, 2011
|
|
3PUN
 
 | | Crystal structure of P domain dimer of Norovirus VA207 with Lewis y tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Chen, Y, Tan, M, Xia, M, Hao, N, Zhang, X.C, Huang, P, Jiang, X, Li, X, Rao, Z. | | Deposit date: | 2010-12-06 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens
Plos Pathog., 7, 2011
|
|
4QWQ
 
 | | Crystal structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus | | Descriptor: | Response regulator SaeR | | Authors: | Fan, X, Zhu, Y, Zhang, X, Teng, M, Li, X. | | Deposit date: | 2014-07-17 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5Z3L
 
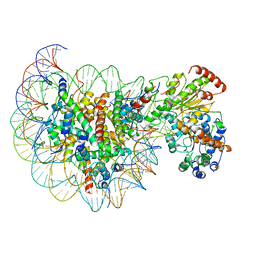 | | Structure of Snf2-nucleosome complex in apo state | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5Z3V
 
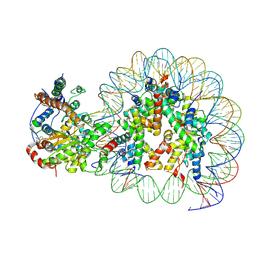 | | Structure of Snf2-nucleosome complex at shl-2 in ADP BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5Z3O
 
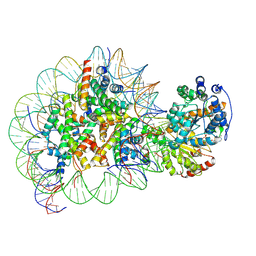 | | Structure of Snf2-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (167-MER), Histone H2A, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5Z3U
 
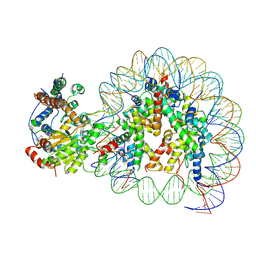 | | Structure of Snf2-nucleosome complex at shl2 in ADP BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
6OHT
 
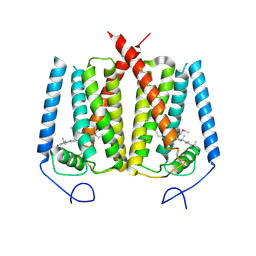 | | Structure of EBP and U18666A | | Descriptor: | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase, 3beta-(2-Diethylaminoethoxy)androst-5-en-17-one | | Authors: | Long, T, Li, X. | | Deposit date: | 2019-04-06 | | Release date: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for human sterol isomerase in cholesterol biosynthesis and multidrug recognition.
Nat Commun, 10, 2019
|
|
7SHN
 
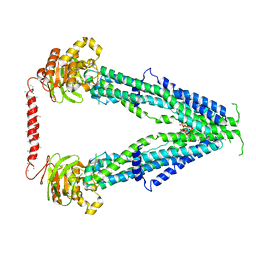 | | Cryo-EM structure of oleoyl-CoA-bound human peroxisomal fatty acid transporter ABCD1 | | Descriptor: | ATP-binding cassette sub-family D member 1, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | Authors: | Wang, R, Li, X. | | Deposit date: | 2021-10-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of acyl-CoA transport across the peroxisomal membrane by human ABCD1.
Cell Res., 32, 2022
|
|
7SHM
 
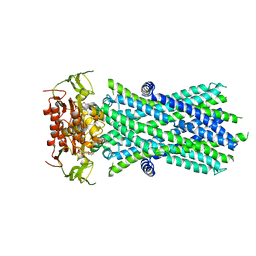 | |
6OHU
 
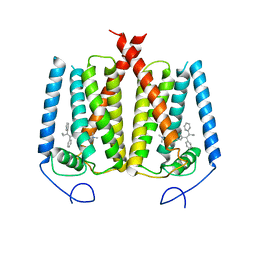 | | Structure of EBP and tamoxifen | | Descriptor: | (Z)-2-[4-(1,2)-DIPHENYL-1-BUTENYL)-PHENOXY]-N,N-DIMETHYLETHANAMINE, 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | | Authors: | Long, T, Li, X. | | Deposit date: | 2019-04-06 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.526 Å) | | Cite: | Structural basis for human sterol isomerase in cholesterol biosynthesis and multidrug recognition.
Nat Commun, 10, 2019
|
|
6OYY
 
 | | Crystal structure of Mtb aspartate decarboxylase, pyrazinoic acid complex | | Descriptor: | Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain, PYRAZINE-2-CARBOXYLIC ACID | | Authors: | Sun, Q, Li, X, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-05-15 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The molecular basis of pyrazinamide activity on Mycobacterium tuberculosis PanD.
Nat Commun, 11, 2020
|
|
6P02
 
 | | Crystal structure of Mtb aspartate decarboxylase, 6-Chlorine pyrazinoic acid complex | | Descriptor: | 6-chloropyrazine-2-carboxylic acid, Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain | | Authors: | Sun, Q, Li, X, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-05-16 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The molecular basis of pyrazinamide activity on Mycobacterium tuberculosis PanD.
Nat Commun, 11, 2020
|
|
4RDJ
 
 | | Crystal structure of Norovirus Boxer P domain | | Descriptor: | Capsid | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
4RDL
 
 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis y tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
7TBW
 
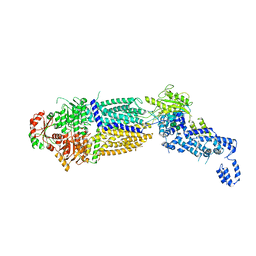 | | The structure of ATP-bound ABCA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Sun, Y, Li, X. | | Deposit date: | 2021-12-22 | | Release date: | 2022-01-26 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cholesterol efflux mechanism revealed by structural analysis of human ABCA1 conformational states.
Nat Cardiovasc Res, 1, 2022
|
|
7TC0
 
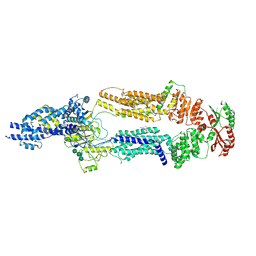 | | The structure of human ABCA1 in digitonin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP-binding cassette, ... | | Authors: | Sun, Y, Li, X. | | Deposit date: | 2021-12-22 | | Release date: | 2022-01-26 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cholesterol efflux mechanism revealed by structural analysis of human ABCA1 conformational states.
Nat Cardiovasc Res, 1, 2022
|
|
7TBY
 
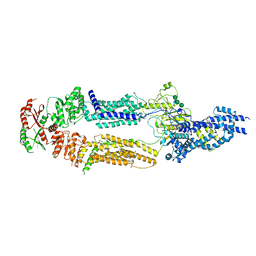 | | The structure of human ABCA1 in nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP-binding cassette, ... | | Authors: | Sun, Y, Li, X. | | Deposit date: | 2021-12-22 | | Release date: | 2022-01-26 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cholesterol efflux mechanism revealed by structural analysis of human ABCA1 conformational states.
Nat Cardiovasc Res, 1, 2022
|
|
4RDK
 
 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis b tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
7TBZ
 
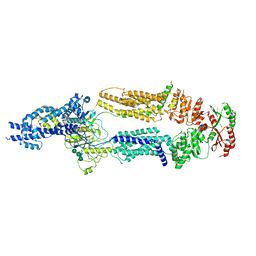 | | The structure of ABCA1 Y482C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP-binding cassette, ... | | Authors: | Sun, Y, Li, X. | | Deposit date: | 2021-12-22 | | Release date: | 2022-01-26 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cholesterol efflux mechanism revealed by structural analysis of human ABCA1 conformational states.
Nat Cardiovasc Res, 1, 2022
|
|
6OZ8
 
 | |
6P1Y
 
 | | Crystal structure of Mtb aspartate decarboxylase mutant M117I | | Descriptor: | AMMONIUM ION, Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain, ... | | Authors: | Sun, Q, Li, X, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-05-20 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The molecular basis of pyrazinamide activity on Mycobacterium tuberculosis PanD.
Nat Commun, 11, 2020
|
|
6PSL
 
 | | Structure of a N-Me-D-Gln4,D-aza-Thr8,Arg10-teixobactin analogue | | Descriptor: | CHLORIDE ION, teixobactin analogue | | Authors: | Nowick, J.S, Yang, H, Pishenko, A, Li, X. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Study of Lactam and Ring-Expanded Analogues of Teixobactin.
J.Org.Chem., 85, 2020
|
|
