2EAV
 
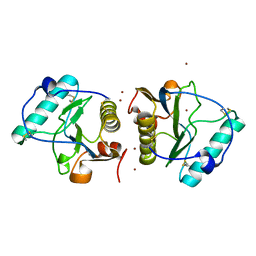 | |
2EAX
 
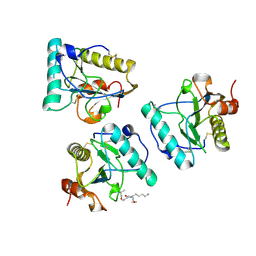 | | Crystal structure of human PGRP-IBETAC in complex with glycosamyl muramyl pentapeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, GLYCOSAMYL MURAMYL PENTAPEPTIDE, Peptidoglycan recognition protein-I-beta | | Authors: | Cho, S. | | Deposit date: | 2007-02-03 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6MH6
 
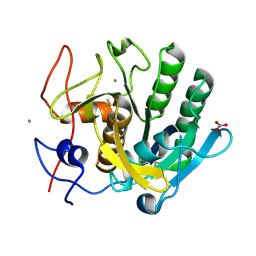 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source. | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
4CJN
 
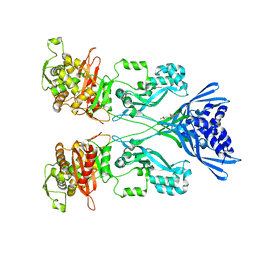 | | Crystal structure of PBP2a from MRSA in complex with quinazolinone ligand | | Descriptor: | (E)-3-(2-(4-cyanostyryl)-4-oxoquinazolin-3(4H)-yl)benzoic acid, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Bouley, R, Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2013-12-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Discovery of Antibiotic (E)-3-(3-Carboxyphenyl)-2-(4-Cyanostyryl)Quinazolin-4(3H)-One.
J.Am.Chem.Soc., 137, 2015
|
|
5CTV
 
 | | Catalytic domain of LytA, the major autolysin of Streptococcus pneumoniae, (C60A, H133A, C136A mutant) complexed with peptidoglycan fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, Autolysin, fragment of peptidoglycan | | Authors: | Achour, A, Sandalova, T, Mellroth, P. | | Deposit date: | 2015-07-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystal structure of the major pneumococcal autolysin LytA in complex with a large peptidoglycan fragment reveals the pivotal role of glycans for lytic activity.
Mol.Microbiol., 101, 2016
|
|
7LAM
 
 | | Crystal structure of Campylobacter jejuni Cj0843c lytic transglycosylase in complex with N,N',N''-triacetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CITRIC ACID, ... | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2021-01-06 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Turnover Chemistry and Structural Characterization of the Cj0843c Lytic Transglycosylase of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
7LAQ
 
 | |
5CA5
 
 | | Structure of the C. elegans NONO-1 homodimer | | Descriptor: | 1,2-ETHANEDIOL, NONO-1 | | Authors: | Knott, G.J, Bond, C.S. | | Deposit date: | 2015-06-29 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Caenorhabditis elegans NONO-1: Insights into DBHS protein structure, architecture, and function.
Protein Sci., 24, 2015
|
|
3U6J
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a pyrazolone inhibitor | | Descriptor: | N-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
3U6I
 
 | | Crystal structure of c-Met in complex with pyrazolone inhibitor 58a | | Descriptor: | Hepatocyte growth factor receptor, N-{3-fluoro-4-[(7-methoxyquinolin-4-yl)oxy]phenyl}-1-[(2R)-2-hydroxypropyl]-5-methyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
4OZS
 
 | | RNA binding protein | | Descriptor: | Alpha solenoid protein | | Authors: | Gully, B.S, Bond, C.S. | | Deposit date: | 2014-02-19 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The design and structural characterization of a synthetic pentatricopeptide repeat protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3U6H
 
 | | Crystal structure of c-Met in complex with pyrazolone inhibitor 26 | | Descriptor: | Hepatocyte growth factor receptor, N-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
2HJ4
 
 | |
2HJB
 
 | |
1C29
 
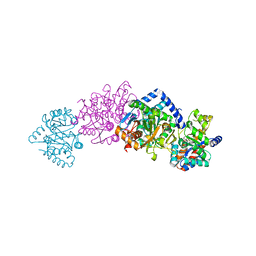 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID | | Descriptor: | 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-07-23 | | Release date: | 2000-01-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
1C9D
 
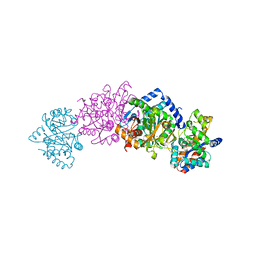 | |
1C8V
 
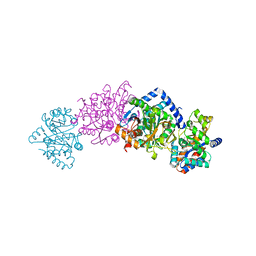 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-HYDROXYPHENYLTHIO)-BUTYLPHOSPHONIC ACID | | Descriptor: | 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-07-30 | | Release date: | 2000-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
2Q7Q
 
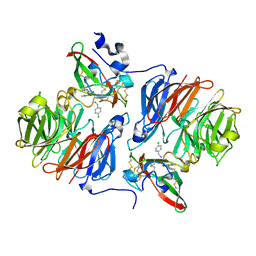 | |
6KN5
 
 | |
6LAG
 
 | | Solution structure of SPA-2 SHD | | Descriptor: | Spa2-like protein | | Authors: | Fan, J.S, Wong, J.Y, Zheng, P, Yang, D, Jedd, G. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spitzenkorper assembly mechanisms reveal conserved features of fungal and metazoan polarity scaffolds.
Nat Commun, 11, 2020
|
|
7KEQ
 
 | | avibactam-CDD-1 6 minute complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (2S,5R)-7-oxo-6-(sulfooxy)-1,6-diazabicyclo[3.2.1]octane-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-01-20 | | Last modified: | 2021-05-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of the Clostridioides difficile Class D beta-Lactamase CDD-1 by Avibactam.
Acs Infect Dis., 7, 2021
|
|
1CW2
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-HYDROXYPHENYLSULFINYL)-BUTYLPHOSPHONIC ACID | | Descriptor: | 4-(2-HYDROXYPHENYLSULFINYL)-BUTYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-08-25 | | Release date: | 1999-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
7JVU
 
 | | Crystal structure of human histone deacetylase 8 (HDAC8) I45T mutation complexed with SAHA | | Descriptor: | 1,2-ETHANEDIOL, Histone deacetylase 8, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, ... | | Authors: | Osko, J.D, Christianson, D.W, Decroos, C, Porter, N.J, Lee, M. | | Deposit date: | 2020-08-24 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5004766 Å) | | Cite: | Structural analysis of histone deacetylase 8 mutants associated with Cornelia de Lange Syndrome spectrum disorders.
J.Struct.Biol., 213, 2020
|
|
7JVW
 
 | | Crystal structure of human histone deacetylase 8 (HDAC8) G320R mutation complexed with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Osko, J.D, Christianson, D.W, Decroos, C, Porter, N.J, Lee, M. | | Deposit date: | 2020-08-24 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.40301776 Å) | | Cite: | Structural analysis of histone deacetylase 8 mutants associated with Cornelia de Lange Syndrome spectrum disorders.
J.Struct.Biol., 213, 2020
|
|
7JVV
 
 | | Crystal structure of human histone deacetylase 8 (HDAC8) E66D/Y306F double mutation complexed with a tetrapeptide substrate | | Descriptor: | 1,2-ETHANEDIOL, ACE-ARG-HIS-ALY-ALY-MCM, GLYCEROL, ... | | Authors: | Osko, J.D, Christianson, D.W, Decroos, C, Porter, N.J, Lee, M. | | Deposit date: | 2020-08-24 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of histone deacetylase 8 mutants associated with Cornelia de Lange Syndrome spectrum disorders.
J.Struct.Biol., 213, 2020
|
|
