2OFS
 
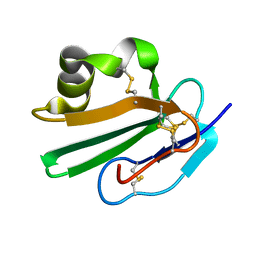 | | Crystal structure of human CD59 | | Descriptor: | CD59 glycoprotein | | Authors: | Davies, C. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of CD59: implications for molecular recognition of the complement proteins C8 and C9 in the membrane-attack complex.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
8H0F
 
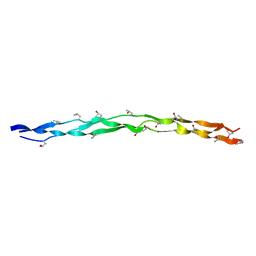 | | Crystal structure of collagen heterotrimer with KD,ER and KE axial pairs | | Descriptor: | collagen-like peptide chain A, collagen-like peptide chain B, collagen-like peptide chain C | | Authors: | Fan, S. | | Deposit date: | 2022-09-28 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Stability of collagen heterotrimer with same charge pattern and different charged residue identities.
Biophys.J., 122, 2023
|
|
8GZO
 
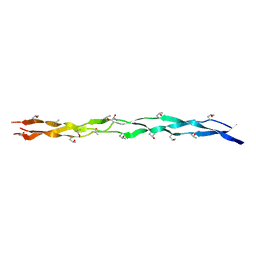 | |
8H0E
 
 | | Crystal structure of collagen heterotrimer with KD, ER and KE axial pairs | | Descriptor: | collagen-like peptide chain A, collagen-like peptide chain B, collagen-like peptide chain C | | Authors: | Fan, S. | | Deposit date: | 2022-09-28 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Stability of collagen heterotrimer with same charge pattern and different charged residue identities.
Biophys.J., 122, 2023
|
|
5JDK
 
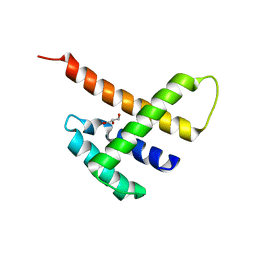 | |
6XC4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 heavy chain, CC12.3 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6XC3
 
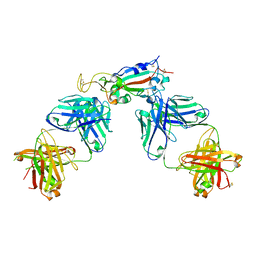 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.1 and CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.1 heavy chain, CC12.1 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6XC7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.3 and CR3022 | | Descriptor: | CC12.3 heavy chain, CC12.3 light chain, CR3022 heavy chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6XC2
 
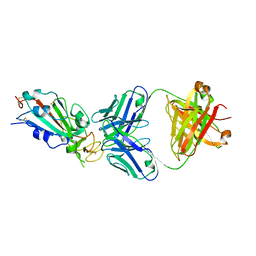 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.1 heavy chain, CC12.1 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.112 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
8JGR
 
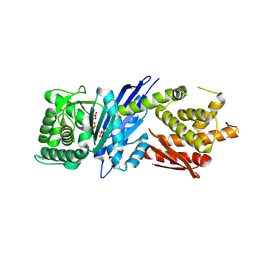 | |
8JGT
 
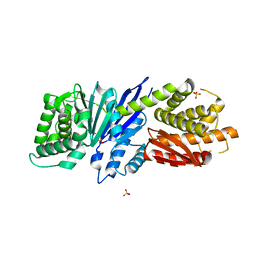 | |
8JGX
 
 | |
8JGW
 
 | |
8JGU
 
 | |
8JGQ
 
 | |
8JGP
 
 | |
8JGO
 
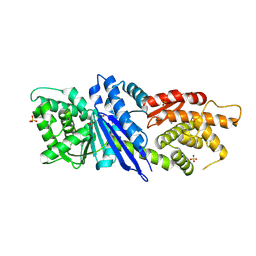 | |
8SLN
 
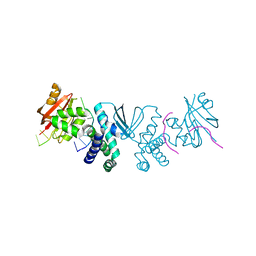 | |
8SLM
 
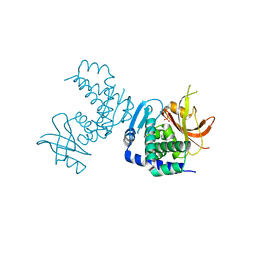 | | Crystal structure of Deinococcus geothermalis PprI | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Zn dependent hydrolase fused to HTH domain, ... | | Authors: | Zhao, Y, Lu, H. | | Deposit date: | 2023-04-23 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The Deinococcus protease PprI senses DNA damage by directly interacting with single-stranded DNA.
Nat Commun, 15, 2024
|
|
4DL7
 
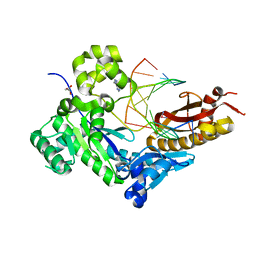 | | Human DNA polymerase eta fails to extend primer 2 nucleotide after cisplatin crosslink (Pt-GG4). | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Cisplatin, DNA (5'-D(*TP*AP*CP*TP*CP*GP*GP*TP*CP*AP*CP*T)-3'), ... | | Authors: | Zhao, Y, Biertumpfel, C, Gregory, M, Hua, Y, Hanaoka, F, Yang, W. | | Deposit date: | 2012-02-05 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis for Chemoresistance to Cisplatin Mediated by DNA Polymerase eta
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DL2
 
 | | Human DNA polymerase eta inserting dCMPNPP opposite CG template (GG0a) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*CP*GP*GP*CP*TP*CP*AP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*TP*GP*AP*G)-3'), ... | | Authors: | Zhao, Y, Biertumpfel, C, Gregory, M, Hua, Y, Hanaoka, F, Yang, W. | | Deposit date: | 2012-02-05 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Chemoresistance to Cisplatin Mediated by DNA Polymerase eta
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DL6
 
 | | Human DNA polymerase eta extending primer immediately after cisplatin crosslink (Pt-GG3). | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, Cisplatin, DNA (5'-D(*TP*AP*GP*TP*GP*TP*GP*CP*C)-3'), ... | | Authors: | Zhao, Y, Biertumpfel, C, Gregory, M, Hua, Y, Hanaoka, F, Yang, W. | | Deposit date: | 2012-02-05 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Chemoresistance to Cisplatin Mediated by DNA Polymerase eta
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DL3
 
 | | Human DNA polymerase eta inserting dCMPNPP opposite GG template (GG0b). | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*TP*AP*CP*GP*GP*TP*CP*AP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*TP*GP*AP*C)-3'), ... | | Authors: | Zhao, Y, Biertumpfel, C, Gregory, M, Hua, Y, Hanaoka, F, Yang, W. | | Deposit date: | 2012-02-05 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Chemoresistance to Cisplatin Mediated by DNA Polymerase eta
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8H6T
 
 | | Complex structure of CDK2/Cyclin E1 and a potent, selective small molecule inhibitor | | Descriptor: | (1R,3S)-3-{3-[(pyridin-2-yl)amino]-1H-pyrazol-5-yl}cyclopentyl propan-2-ylcarbamate, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
8H6P
 
 | | Complex structure of CDK2/Cyclin E1 and a potent, selective macrocyclic inhibitor | | Descriptor: | (7S,10R)-11-oxa-2,4,5,13,17,23-hexaazatetracyclo[17.3.1.1~3,6~.1~7,10~]pentacosa-1(23),3(25),5,19,21-pentaene-12,18-dione, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
