6MBD
 
 | | Human Mcl-1 in complex with the designed peptide dM1 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION, dM1 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6ME5
 
 | | XFEL crystal structure of human melatonin receptor MT1 in complex with agomelatine | | Descriptor: | OLEIC ACID, chimera protein of Melatonin receptor type 1A and GlgA glycogen synthase, ~{N}-[2-(7-methoxynaphthalen-1-yl)ethyl]ethanamide | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
6MER
 
 | | PcdhgB3 EC1-4 in 50 mM HEPES | | Descriptor: | CALCIUM ION, Protocadherin gamma-B3 | | Authors: | Nicoludis, J.M, Gaudet, R. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interaction specificity of clustered protocadherins inferred from sequence covariation and structural analysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6MFX
 
 | | Crystal structure of a 4-domain construct of a mutant of LgrA in the substrate donation state | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Linear gramicidin synthase subunit A, N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Reimer, J.M, Eivaskhani, M, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6MFZ
 
 | | Crystal structure of dimodular LgrA in a condensation state | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Linear gramicidin synthase subunit A | | Authors: | Reimer, J.M, Eivaskhani, M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6M9U
 
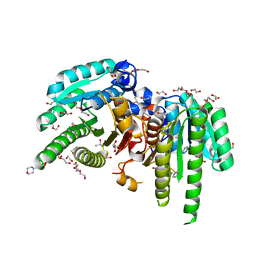 | | Structure of the apo-form of 20beta-Hydroxysteroid Dehydrogenase from Bifidobacterium adolescentis strain L2-32 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Mythen, S.M, Pollet, R.M, Koropatkin, N.M, Ridlon, J.M. | | Deposit date: | 2018-08-24 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of 20 beta-hydroxysteroid dehydrogenase fromBifidobacterium adolescentisstrain L2-32.
J.Biol.Chem., 294, 2019
|
|
6MG0
 
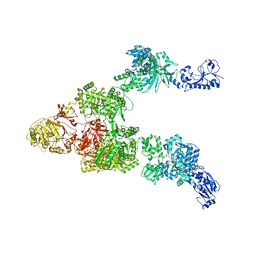 | | Crystal structure of a 5-domain construct of LgrA in the thiolation state | | Descriptor: | 5'-({[(2R,3R)-3-amino-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-{[oxido(oxo)phosphonio]oxy}butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-4-methylpentyl]sulfonyl}amino)-5'-deoxyadenosine, Linear gramicidin synthase subunit A | | Authors: | Reimer, J.M, Eivaskhani, M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6ME3
 
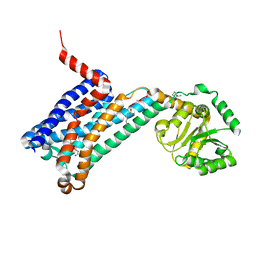 | | XFEL crystal structure of human melatonin receptor MT1 in complex with 2-phenylmelatonin | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, OLEIC ACID, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
6MFW
 
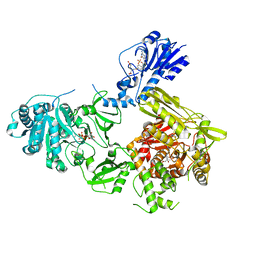 | | Crystal structure of a 4-domain construct of LgrA in the substrate donation state | | Descriptor: | (2~{R})-~{N}-[3-[2-[[(2~{S})-2-formamido-3-methyl-butanoyl]amino]ethylamino]-3-oxidanylidene-propyl]-3,3-dimethyl-2-oxidanyl-4-[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxy-butanamide, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Linear gramicidin synthase subunit A, ... | | Authors: | Reimer, J.M, Eivaskhani, M, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6MH4
 
 | |
6MEQ
 
 | | PcdhgB3 EC1-4 in 50 mM HEPES | | Descriptor: | CALCIUM ION, Protocadherin gamma-B3 | | Authors: | Nicoludis, J.M, Gaudet, R. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interaction specificity of clustered protocadherins inferred from sequence covariation and structural analysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6MBB
 
 | | Human Bfl-1 in complex with the designed peptide dF1 | | Descriptor: | Bcl-2-related protein A1, dF1 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6ME2
 
 | | XFEL crystal structure of human melatonin receptor MT1 in complex with ramelteon | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}propanamide, OLEIC ACID, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
6ME4
 
 | | XFEL crystal structure of human melatonin receptor MT1 in complex with 2-iodomelatonin | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-[2-(2-iodo-5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
6MH6
 
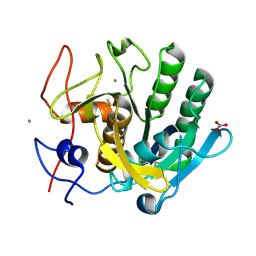 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source. | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
6MNI
 
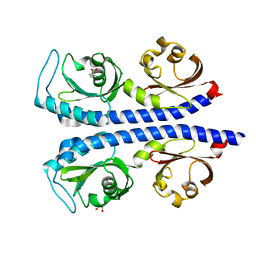 | | Structure of the tandem CACHE domain of PscC | | Descriptor: | CHLORIDE ION, GLYCEROL, Methyl-accepting chemotaxis protein, ... | | Authors: | Johnston, J.M, Gerth, M.L, Ehrhardt, M.K.G. | | Deposit date: | 2018-10-01 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structure of a double CACHE chemoreceptor ligand-binding domain from Pseudomonas syringae provides insights into the basis of proline recognition.
Biochem.Biophys.Res.Commun., 549, 2021
|
|
6MH5
 
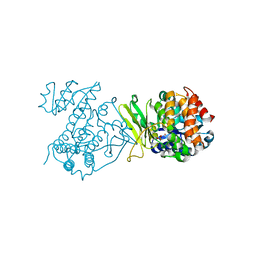 | |
6N4X
 
 | | Metabotropic Glutamate Receptor 5 Apo Form Ligand Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 5 | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6N2A
 
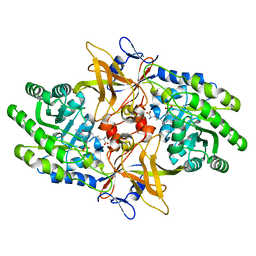 | |
6MH8
 
 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, Soluble cytochrome b562 chimeric construct | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
6K2G
 
 | |
7OOL
 
 | | Crystal structure of a Candidatus photodesmus katoptron thioredoxin chimera containing an ancestral loop | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Gavira, J.A, Ibarra-Molero, B, Gamiz-Arco, G, Risso, V, Sanchez-Ruiz, J.M. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Combining Ancestral Reconstruction with Folding-Landscape Simulations to Engineer Heterologous Protein Expression.
J.Mol.Biol., 433, 2021
|
|
7ON3
 
 | | SaFtsZ complexed with GDP (soak 10 mM EGTA) | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OMP
 
 | | SaFtsZ complexed with GDPPCP and Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, MAGNESIUM ION, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7ON4
 
 | | SaFtsZ complexed with GDP (co-crystalization with 1mM EDTA) | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
