1I7U
 
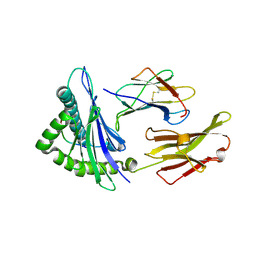 | | CRYSTAL STRUCTURE OF CLASS I MHC A2 IN COMPLEX WITH PEPTIDE P1049-6V | | Descriptor: | 9 RESIDUE PEPTIDE, BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Busslep, J, Zhao, R, Loftus, D, Appella, E, Collins, E.J. | | Deposit date: | 2001-03-10 | | Release date: | 2001-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | T cell activity correlates with oligomeric peptide-major histocompatibility complex binding on T cell surface
J.Biol.Chem., 276, 2001
|
|
9GA3
 
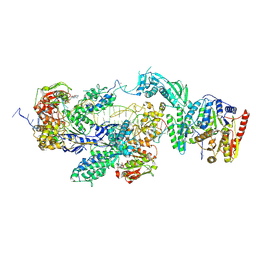 | | MtUvrA2UvrB bound to damaged oligonucleotide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA, UvrABC system protein A, ... | | Authors: | Genta, M, Capelli, R, Ferrara, G, Rizzi, M, Rossi, F, Jeruzalmi, D, Bolognesi, M, Chaves-Sanjuan, A, Miggiano, R. | | Deposit date: | 2024-07-26 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanistic understanding of UvrA damage detection and lesion hand-off to UvrB in Nucleotide Excision Repair.
Nat Commun, 16, 2025
|
|
7UAC
 
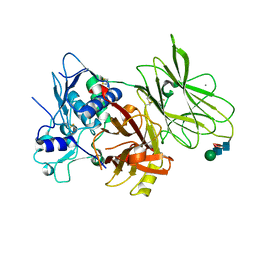 | | Human pro-meprin alpha (zymogen state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Meprin A subunit alpha, ... | | Authors: | Bayly-Jones, C, Lupton, C.J, Fritz, C, Schlenzig, D, Whisstock, J.C. | | Deposit date: | 2022-03-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Helical ultrastructure of the metalloprotease meprin alpha in complex with a small molecule inhibitor.
Nat Commun, 13, 2022
|
|
7UAB
 
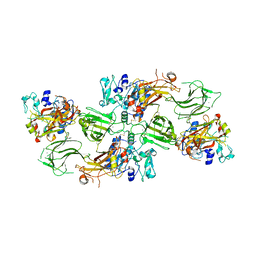 | | Human pro-meprin alpha (zymogen state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bayly-Jones, C, Lupton, C.J, Fritz, C, Schlenzig, D, Whisstock, J.C. | | Deposit date: | 2022-03-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Helical ultrastructure of the metalloprotease meprin alpha in complex with a small molecule inhibitor.
Nat Commun, 13, 2022
|
|
7UAI
 
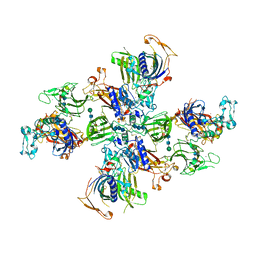 | | Meprin alpha helix in complex with fetuin-B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bayly-Jones, C, Lupton, C.J, Fritz, C, Schlenzig, D, Whisstock, J.C. | | Deposit date: | 2022-03-13 | | Release date: | 2022-11-02 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Helical ultrastructure of the metalloprotease meprin alpha in complex with a small molecule inhibitor.
Nat Commun, 13, 2022
|
|
7UAE
 
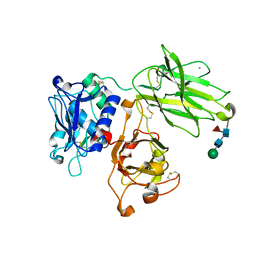 | | Human meprin alpha (active state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bayly-Jones, C, Lupton, C.J, Fritz, C, Schlenzig, D, Whisstock, J.C. | | Deposit date: | 2022-03-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Helical ultrastructure of the metalloprotease meprin alpha in complex with a small molecule inhibitor.
Nat Commun, 13, 2022
|
|
8PIX
 
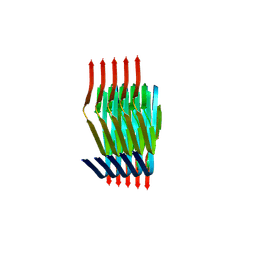 | | Cryo EM structure of the type 3C polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
8PJO
 
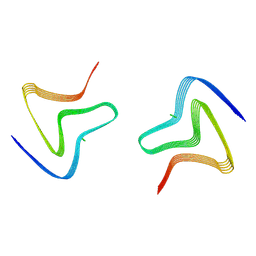 | | Cryo EM structure of the type 3D polymorph of alpha-synuclein E46K mutant at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
9GA5
 
 | | MtUvrA2 bound to endogenous E. coli DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Endogenous E. coli DNA, UvrABC system protein A, ... | | Authors: | Genta, M, Capelli, R, Ferrara, G, Rizzi, M, Rossi, F, Jeruzalmi, D, Bolognesi, M, Chaves-Sanjuan, A, Miggiano, R. | | Deposit date: | 2024-07-26 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanistic understanding of UvrA damage detection and lesion hand-off to UvrB in Nucleotide Excision Repair.
Nat Commun, 16, 2025
|
|
3MGJ
 
 | | Crystal structure of the Saccharop_dh_N domain of MJ1480 protein from Methanococcus jannaschii. Northeast Structural Genomics Consortium Target MjR83a. | | Descriptor: | Uncharacterized protein MJ1480 | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Lee, D, Patel, D, Ciccosanti, C, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-06 | | Release date: | 2010-04-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structure of the Saccharop_dh_N domain of MJ1480 protein from Methanococcus jannaschii.
To be Published
|
|
1AH2
 
 | | SERINE PROTEASE PB92 FROM BACILLUS ALCALOPHILUS, NMR, 18 STRUCTURES | | Descriptor: | SERINE PROTEASE PB92 | | Authors: | Boelens, R, Schipper, D, Martin, J.R, Karimi-Nejad, Y, Mulder, F, Zwan, J.V.D, Mariani, M. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of serine protease PB92 from Bacillus alcalophilus presents a rigid fold with a flexible substrate-binding site.
Structure, 5, 1997
|
|
1L9L
 
 | | GRANULYSIN FROM HUMAN CYTOLYTIC T LYMPHOCYTES | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ETHANOL, Granulysin, ... | | Authors: | Anderson, D.H, Sawaya, M.R, Cascio, D, Ernst, W, Krensky, A, Modlin, R, Eisenberg, D. | | Deposit date: | 2002-03-25 | | Release date: | 2002-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Granulysin Crystal Structure and a Structure-Derived Lytic Mechanism
J.Mol.Biol., 325, 2002
|
|
1ADV
 
 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C, Tucker, P.A. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
1AAM
 
 | | THE STRUCTURAL BASIS FOR THE ALTERED SUBSTRATE SPECIFICITY OF THE R292D ACTIVE SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | Aspartate aminotransferase, SULFATE ION | | Authors: | Almo, S.C, Smith, D.L, Danishefsky, A.T, Ringe, D. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for the altered substrate specificity of the R292D active site mutant of aspartate aminotransferase from E. coli.
Protein Eng., 7, 1994
|
|
9GC6
 
 | | Highly optimized CNS penetrant inhibitors of EGFR Exon20 Insertion Mutations | | Descriptor: | 1-[2-[5-fluoranyl-4-(2-fluorophenyl)pyridin-2-yl]-3-pyrimidin-4-yl-4,6-dihydropyrrolo[3,4-d]imidazol-5-yl]propan-1-one, Epidermal growth factor receptor | | Authors: | Hargreaves, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Highly Optimized CNS Penetrant Inhibitors of EGFR Exon20 Insertion Mutations.
J.Med.Chem., 68, 2025
|
|
9GC5
 
 | | Highly optimized CNS penetrant inhibitors of EGFR Exon20 Insertion Mutations | | Descriptor: | 1-[2-[5-(1,3-benzoxazol-4-yl)-2,4-bis(fluoranyl)phenyl]-3-pyrimidin-4-yl-4,6-dihydropyrrolo[3,4-d]imidazol-5-yl]propan-1-one, Epidermal growth factor receptor, SULFATE ION | | Authors: | Hargreaves, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | Highly Optimized CNS Penetrant Inhibitors of EGFR Exon20 Insertion Mutations.
J.Med.Chem., 68, 2025
|
|
9GC4
 
 | | Highly optimized CNS penetrant inhibitors of EGFR Exon20 Insertion Mutations | | Descriptor: | 1-[2-[3-(3-chloranyl-6-fluoranyl-pyridin-2-yl)oxyphenyl]-3-pyrimidin-4-yl-4,6-dihydropyrrolo[3,4-d]imidazol-5-yl]propan-1-one, Epidermal growth factor receptor | | Authors: | Hargreaves, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | Highly Optimized CNS Penetrant Inhibitors of EGFR Exon20 Insertion Mutations.
J.Med.Chem., 68, 2025
|
|
9GDV
 
 | | Highly optimized CNS penetrant inhibitors of EGFR Exon20 Insertion Mutations | | Descriptor: | Epidermal growth factor receptor, SULFATE ION, ~{N}-[2-[2-(dimethylamino)ethyl-methyl-amino]-4-methoxy-5-[[4-(1-methylindol-3-yl)pyrimidin-2-yl]amino]phenyl]propanamide | | Authors: | Hargreaves, D. | | Deposit date: | 2024-08-06 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | Highly Optimized CNS Penetrant Inhibitors of EGFR Exon20 Insertion Mutations.
J.Med.Chem., 68, 2025
|
|
5FGO
 
 | |
1NNO
 
 | | CONFORMATIONAL CHANGES OCCURRING UPON NO BINDING IN NITRITE REDUCTASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, NITRIC OXIDE, ... | | Authors: | Nurizzo, D, Tegoni, M, Cambillau, C. | | Deposit date: | 1998-07-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Conformational changes occurring upon reduction and NO binding in nitrite reductase from Pseudomonas aeruginosa.
Biochemistry, 37, 1998
|
|
7U4W
 
 | | The ubiquitin-associated domain of human thirty-eight negative kinase-1 flexibly fused to the 1TEL crystallization chaperone via a 2-glycine linker and crystallized at traditional protein concentration | | Descriptor: | Transcription factor ETV6,Non-receptor tyrosine-protein kinase TNK1 | | Authors: | Nawarathnage, S, Pedroza Romo, M.J, Smith, T, Bunn, D, Stewart, C, Doukov, T, Moody, J.D. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fusion crystallization reveals the behavior of both the 1TEL crystallization chaperone and the TNK1 UBA domain.
Structure, 31, 2023
|
|
3MML
 
 | | Allophanate Hydrolase Complex from Mycobacterium smegmatis, Msmeg0435-Msmeg0436 | | Descriptor: | Allophanate hydrolase subunit 1, Allophanate hydrolase subunit 2, CHLORIDE ION | | Authors: | Kaufmann, M, Chernishof, I, Shin, A, Germano, D, Sawaya, M.R, Waldo, G.S, Arbing, M.A, Perry, J, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Allphanate Hydrolase Complex from M. smegmatis, Msmeg0435-Msmeg0436
To be Published
|
|
8F6N
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Thymine Quasi-Anaerobically | | Descriptor: | Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kaley, N, Smith, M, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2022-11-16 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Mammalian dihydropyrimidine dehydrogenase: Added mechanistic details from transient-state analysis of charge transfer complexes.
Arch.Biochem.Biophys., 736, 2023
|
|
8F5W
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Dihydrothymine and NADPH Quasi-Anaerobically | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kaley, N, Smith, M, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-01 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mammalian dihydropyrimidine dehydrogenase: Added mechanistic details from transient-state analysis of charge transfer complexes.
Arch.Biochem.Biophys., 736, 2023
|
|
1JUF
 
 | | Structure of Minor Histocompatibility Antigen peptide, H13b, complexed to H2-Db | | Descriptor: | Beta-2-microglobulin, H13b peptide, H2-Db major histocompatibility antigen | | Authors: | Ostrov, D.A, Roden, M.M, Shi, W, Palmieri, E, Christianson, G.J, Mendoza, L, Villaflor, G, Tilley, D, Shastri, N, Grey, H, Almo, S.C, Roopenian, D.C, Nathenson, S.G. | | Deposit date: | 2001-08-24 | | Release date: | 2002-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How H13 histocompatibility peptides differing by a single methyl group and lacking conventional MHC binding anchor motifs determine self-nonself discrimination.
J.Immunol., 168, 2002
|
|
