7YW2
 
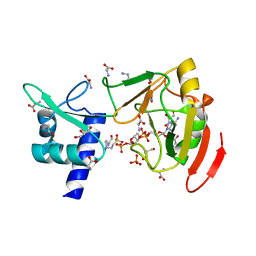 | | Crystal structure of tRNA 2'-phosphotransferase from Mus musculus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, GLYCINE, ... | | Authors: | Yang, X.Y, Liu, X.H. | | Deposit date: | 2022-08-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical insights into the molecular mechanism of TRPT1 for nucleic acid ADP-ribosylation.
Nucleic Acids Res., 51, 2023
|
|
7YW3
 
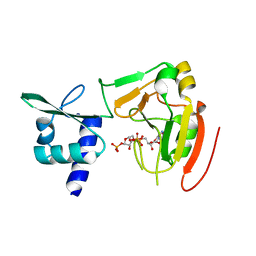 | | Crystal structure of tRNA 2'-phosphotransferase from Homo sapiens | | Descriptor: | 1,2-ETHANEDIOL, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate, tRNA 2'-phosphotransferase 1 | | Authors: | Yang, X.Y, Liu, X.H. | | Deposit date: | 2022-08-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical insights into the molecular mechanism of TRPT1 for nucleic acid ADP-ribosylation.
Nucleic Acids Res., 51, 2023
|
|
7YW4
 
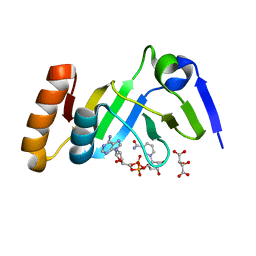 | |
5YUD
 
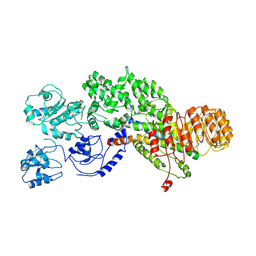 | | Flagellin derivative in complex with the NLR protein NAIP5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Baculoviral IAP repeat-containing protein 1e, Phase 2 flagellin,Flagellin | | Authors: | Yang, X.R, Yang, F, Wang, W.G, Lin, G.Z. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-03 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structural basis for specific flagellin recognition by the NLR protein NAIP5.
Cell Res., 28, 2018
|
|
8JFL
 
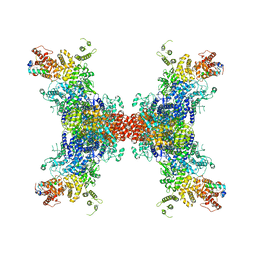 | | PhK holoenzyme in active state, muscle isoform | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FARNESYL, Phosphorylase b kinase gamma catalytic chain, ... | | Authors: | Yang, X.K, Xiao, J.Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture and activation of human muscle phosphorylase kinase.
Nat Commun, 15, 2024
|
|
8JFK
 
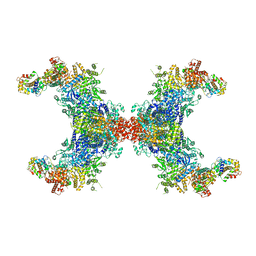 | | PhK holoenzyme in inactive state, muscle isoform | | Descriptor: | Calmodulin-1, FARNESYL, Phosphorylase b kinase gamma catalytic chain, ... | | Authors: | Yang, X.K, Xiao, J.Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture and activation of human muscle phosphorylase kinase.
Nat Commun, 15, 2024
|
|
3AJF
 
 | |
5YTX
 
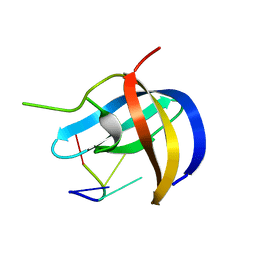 | | Crystal structure of YB1 cold-shock domain in complex with UCAACU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*UP*CP*AP*AP*CP*U)-3') | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
5YTT
 
 | | Crystal structure of YB1 cold-shock domain in complex with UCAUGU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*UP*CP*AP*UP*GP*U)-3'), SULFATE ION | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
5YTV
 
 | | Crystal structure of YB1 cold-shock domain in complex with UCAUCU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*UP*CP*AP*UP*CP*U)-3') | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
5YTS
 
 | | Crystal structure of YB1 cold-shock domain in complex with UCUUCU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*CP*UP*UP*C)-3'), SULFATE ION | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
8XYB
 
 | | hPhK gamma-delta subcomplex in inactive state | | Descriptor: | Calmodulin-1, FARNESYL, Phosphorylase b kinase gamma catalytic chain, ... | | Authors: | Yang, X.K, Xiao, J.Y. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Architecture and activation of human muscle phosphorylase kinase.
Nat Commun, 15, 2024
|
|
8XY7
 
 | | hPhK alpha-gamma subcomplex in active state | | Descriptor: | FARNESYL, Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform, ... | | Authors: | Yang, X.K, Xiao, J.Y. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture and activation of human muscle phosphorylase kinase.
Nat Commun, 15, 2024
|
|
8XYA
 
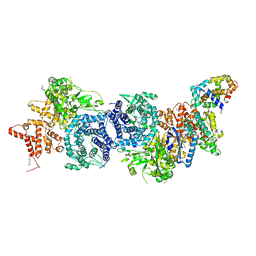 | |
5ZOL
 
 | | Crystal structure of a three sites mutantion of FSAA complexed with HA and product | | Descriptor: | (3S,4S)-3,4-dihydroxy-4-(thiophen-2-yl)butan-2-one, 1-hydroxypropan-2-one, CHLORIDE ION, ... | | Authors: | Wu, L, Yang, X.H, Yu, H.W, Zhou, J.H. | | Deposit date: | 2018-04-13 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | The engineering of decameric d-fructose-6-phosphate aldolase A by combinatorial modulation of inter- and intra-subunit interactions.
Chem.Commun.(Camb.), 56, 2020
|
|
7WV9
 
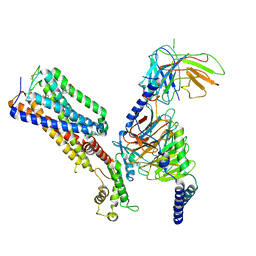 | | Allosteric modulator ZCZ011 binding to CP55940-bound cannabinoid receptor 1 in complex with Gi protein | | Descriptor: | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, 6-methyl-3-[(1S)-2-nitro-1-thiophen-2-yl-ethyl]-2-phenyl-1H-indole, Cannabinoid receptor 1, ... | | Authors: | Xu, Z, Shao, Z. | | Deposit date: | 2022-02-10 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1.
Nat.Chem.Biol., 18, 2022
|
|
7W8N
 
 | | Microbial Hormone-sensitive lipase E53 wild type | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ... | | Authors: | Yang, X, Li, Z, Xu, X, Li, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-02-23 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism and Structural Insights Into a Novel Esterase, E53, Isolated From Erythrobacter longus .
Front Microbiol, 12, 2021
|
|
7CI0
 
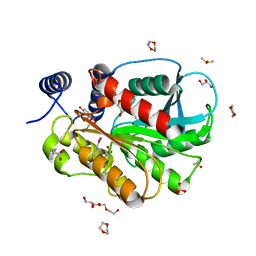 | | Microbial Hormone-sensitive lipase E53 mutant S162A | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Yang, X, Li, Z, Xu, X, Li, J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism and Structural Insights Into a Novel Esterase, E53, Isolated From Erythrobacter longus .
Front Microbiol, 12, 2021
|
|
7CIH
 
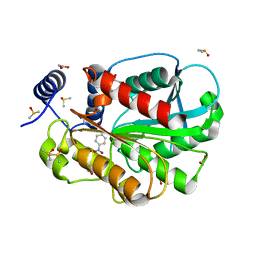 | | Microbial Hormone-sensitive lipase E53 mutant S285G | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ... | | Authors: | Yang, X, Li, Z, Xu, X, Li, J. | | Deposit date: | 2020-07-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Mechanism and Structural Insights Into a Novel Esterase, E53, Isolated From Erythrobacter longus .
Front Microbiol, 12, 2021
|
|
8H3S
 
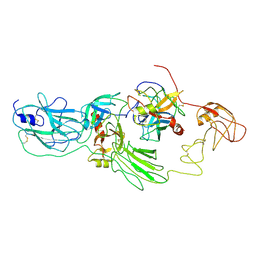 | | Substrate-bound EP, polyA model | | Descriptor: | Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain, Serine protease 1 | | Authors: | Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-10-09 | | Release date: | 2022-11-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
8H3U
 
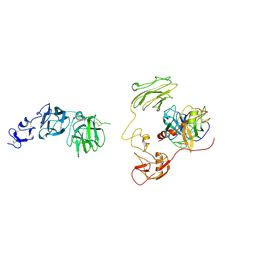 | | Inhibitor-bound EP, polyA model | | Descriptor: | Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain | | Authors: | Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-10-09 | | Release date: | 2022-11-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
7WDK
 
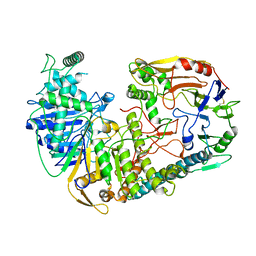 | | The structure of PldA-PA3488 complex | | Descriptor: | Phospholipase D, Tli4_C domain-containing protein | | Authors: | Zhao, L, Yang, X.Y, Li, Z.Q. | | Deposit date: | 2021-12-21 | | Release date: | 2022-10-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into PA3488-mediated inactivation of Pseudomonas aeruginosa PldA
Nat Commun, 13, 2022
|
|
7WQZ
 
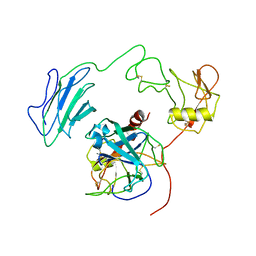 | | Structure of Active-mutEP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain | | Authors: | Yang, X.L, Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-10-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
7WQW
 
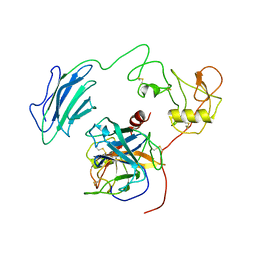 | | Structure of Active-EP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain | | Authors: | Yang, X.L, Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-10-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
7WQX
 
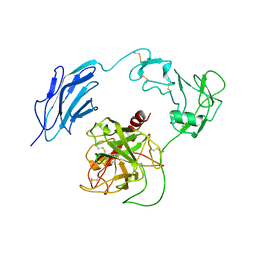 | | Structure of Inactive-EP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Enteropeptidase | | Authors: | Yang, X.L, Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-10-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
