6VI2
 
 | | Structure of the unaligned Fab4 | | Descriptor: | FAB4 heavy chain, FAB4 light chain | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-01-11 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5IX0
 
 | | HDAC2 WITH LIGAND BRD7232 | | Descriptor: | (3-exo)-N-(4-amino-4'-fluoro[1,1'-biphenyl]-3-yl)-8-oxabicyclo[3.2.1]octane-3-carboxamide, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Steinbacher, S. | | Deposit date: | 2016-03-23 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetic and structural insights into the binding of histone deacetylase 1 and 2 (HDAC1, 2) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
7AV4
 
 | | Dark state structure of the C432S mutant of Fatty Acid Photodecarboxylase (FAP) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Schlichting, I, Hartmann, E, Arnoux, P, Sorigue, D, Beisson, F. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
8P2L
 
 | |
8P2M
 
 | | C. elegans TIR-1 protein. | | Descriptor: | NAD(+) hydrolase tir-1 | | Authors: | Isupov, M.N, Opatowsky, Y. | | Deposit date: | 2023-05-16 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure-function analysis of ceTIR-1/hSARM1 explains the lack of Wallerian axonal degeneration in C. elegans.
Cell Rep, 42, 2023
|
|
5IVH
 
 | |
1QHR
 
 | | NOVEL COVALENT ACTIVE SITE THROMBIN INHIBITORS | | Descriptor: | 6-(2-HYDROXY-CYCLOPENTYL)-7-OXO-HEPTANAMIDINE, ALPHA THROMBIN, HIRUGEN | | Authors: | Jhoti, H, Cleasby, A, Reid, S, Thomas, P, Wonacott, A. | | Deposit date: | 1999-05-26 | | Release date: | 2000-05-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of thrombin complexed to a novel series of synthetic inhibitors containing a 5,5-trans-lactone template.
Biochemistry, 38, 1999
|
|
1QJ6
 
 | | Novel Covalent Active Site Thrombin Inhibitors | | Descriptor: | 6-CARBAMIMIDOYL-2-[2-HYDROXY-5-(3-METHOXY-PHENYL)-INDAN-1-YL]-HEXANOIC ACID, HIRUGEN, THROMBIN | | Authors: | Jhoti, H, Cleasby, A. | | Deposit date: | 1999-06-22 | | Release date: | 2000-06-22 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Thrombin Complexed to a Novel Series of Synthetic Inhibitors Containing a 5,5-Trans-Lactone Template
Biochemistry, 38, 1999
|
|
1QJ7
 
 | | Novel Covalent Active Site Thrombin Inhibitors | | Descriptor: | 6-CARBAMIMIDOYL-2-[5-(3-DIETHYLCARBAMOYL-PHENYL)-2-HYDROXY-INDAN-1-YL]-HEXANOIC ACID, HIRUGEN, THROMBIN | | Authors: | Jhoti, H, Cleasby, A. | | Deposit date: | 1999-06-22 | | Release date: | 2000-06-22 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Thrombin Complexed to a Novel Series of Synthetic Inhibitors Containing a 5,5-Trans-Lactone Template
Biochemistry, 38, 1999
|
|
4M6K
 
 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADP+ and folate | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, GLYCEROL, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6GW9
 
 | | Concanavalin A structure determined with data from the EuXFEL, the first MHz free electron laser | | Descriptor: | CALCIUM ION, Concanavalin V, MAGNESIUM ION | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6H0K
 
 | | Hen egg-white lysozyme structure determined with data from the EuXFEL, the first MHz free electron laser, 7.47 keV photon energy | | Descriptor: | Lysozyme C | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6H0L
 
 | | Hen egg-white lysozyme structure determined with data from the EuXFEL, 9.22 keV photon energy | | Descriptor: | Lysozyme C | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6GWA
 
 | | Concanavalin B structure determined with data from the EuXFEL, the first MHz free electron laser | | Descriptor: | Concanavalin B | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
7NCX
 
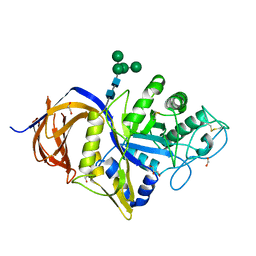 | | Crystal structure of GH30 (double mutant EE) from Thermothelomyces thermophila. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dimarogona, M, Nikolaivits, E, Topakas, E, Weiss, M, Feiler, C.G. | | Deposit date: | 2021-01-29 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Unique features of the bifunctional GH30 from Thermothelomyces thermophila revealed by structural and mutational studies
Carbohydrate Polymers, 273, 2021
|
|
6YFY
 
 | | Solid-state NMR structure of the D-Arg4,L10-teixobactin - Lipid II complex in lipid bilayers. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, 3-methylbut-2-en-1-ol, D-Arg4,Leu10-Teixobactin, ... | | Authors: | Weingarth, M.H, Shukla, R. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-10 | | Last modified: | 2024-08-07 | | Method: | SOLID-STATE NMR | | Cite: | Mode of action of teixobactins in cellular membranes.
Nat Commun, 11, 2020
|
|
7QGV
 
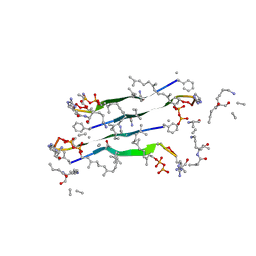 | | Solid-state NMR structure of Teixobactin-Lipid II. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, 3-methylbut-2-en-1-ol, Lipid II, ... | | Authors: | Weingarth, M.H, Shukla, R. | | Deposit date: | 2021-12-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-15 | | Method: | SOLID-STATE NMR | | Cite: | Teixobactin kills bacteria by a two-pronged attack on the cell envelope.
Nature, 608, 2022
|
|
4MMK
 
 | | Q8A Hfq from Pseudomonas aeruginosa | | Descriptor: | POTASSIUM ION, Protein hfq, SODIUM ION, ... | | Authors: | Murina, V.N, Filimonov, V.V, Melnik, B.S, Uhlein, M, Mueller, U, Weiss, M, Nikulin, A.D. | | Deposit date: | 2013-09-09 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Effect of conserved intersubunit amino Acid substitutions on hfq protein structure and stability.
Biochemistry Mosc., 79, 2014
|
|
6YRV
 
 | | Crystal structure of FAP after illumination at 100K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRU
 
 | | Crystal structure of FAP in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YS2
 
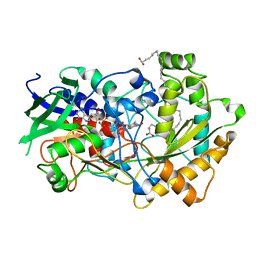 | | Crystal structure of FAP R451A in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRX
 
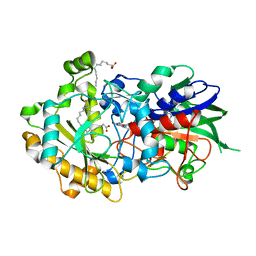 | | Low-dose crystal structure of FAP at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRZ
 
 | | Crystal structure of FAP et pH 8.5 after illumination at 150K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, ... | | Authors: | Sorigue, D, Legrand, P, Blangy, S, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YS1
 
 | | Crystal structure of FAP R451K mutant in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
8R94
 
 | |
