6BRR
 
 | |
5F1R
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (C10) | | Descriptor: | (3~{R})-8-cyclopropyl-7-(naphthalen-1-ylmethyl)-5-oxidanylidene-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, Listeriolysin regulatory protein | | Authors: | Begum, A, Grundstrom, C, Good, J.A.D, Andersson, C, ALmqvist, F, Johansson, J, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Attenuating Listeria monocytogenes Virulence by Targeting the Regulatory Protein PrfA.
Cell Chem Biol, 23, 2016
|
|
5URF
 
 | | The structure of human bocavirus 1 | | Descriptor: | viral protein 3 | | Authors: | Mietzsch, M, Kailasan, S, Garrison, J, Ilyas, M, Chipman, P, Kandola, K, Jansen, M, Spear, J, Sousa, D, McKenna, R, Soderlund-Venermo, M, Baker, T, Agbandje-McKenna, M. | | Deposit date: | 2017-02-10 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Insights into Human Bocaparvoviruses.
J. Virol., 91, 2017
|
|
2WD1
 
 | | Human c-Met Kinase in complex with azaindole inhibitor | | Descriptor: | 1-[(2-NITROPHENYL)SULFONYL]-1H-PYRROLO[3,2-B]PYRIDINE-6-CARBOXAMIDE, GAMMA-BUTYROLACTONE, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | Porter, J, Lumb, S, Franklin, R.J, Gascon-Simorte, J.M, Calmiano, M, Le Riche, K, Lallemand, B, Keyaerts, J, Edwards, H, Maloney, A, Delgado, J, King, L, Foley, A, Lecomte, F, Reuberson, J, Meier, C, Batchelor, M. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-Azaindoles as Novel Inhibitors of C- met Kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4BUM
 
 | |
7DEA
 
 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck Northern China/22/2017 (H5N6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Sun, H, Sun, H, Song, J, Zhang, W, Wei, X, Qi, J, Gao, G.F, Liu, J. | | Deposit date: | 2020-11-03 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
7DEB
 
 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck/Eastern China/L0230/2010 (H5N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Sun, H, Sun, H, Song, J, Zhang, W, Qi, J, Gao, G.F, Liu, J. | | Deposit date: | 2020-11-03 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
2QVK
 
 | |
2QVM
 
 | | The second Ca2+-binding domain of the Na+-Ca2+ exchanger is essential for regulation: crystal structures and mutational analysis | | Descriptor: | CALCIUM ION, Sodium/calcium exchanger 1 | | Authors: | Chaptal, V, Mercado Besserer, G, Abramson, J, Cascio, D. | | Deposit date: | 2007-08-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | The second Ca2+-binding domain of the Na+ Ca2+ exchanger is essential for regulation: crystal structures and mutational analysis
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5UG3
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN GID MUTANT A10V | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A.K, Leffler, A.E, Zebroski, H.A, Powell, S.R, Kuryatov, A, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UG5
 
 | | NMR SOLUTION STRUCTURE OF THE ALPHA-CONOTOXIN GID MUTANT V13Y | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A, Leffler, A.E, Kuryatov, A, Zebroski, H.A, Powell, S.R, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1RNA
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF AN RNA HELIX: [U(U-A)6A]2 | | Descriptor: | RNA (5'-R(*UP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*A)-3') | | Authors: | Dock-Bregeon, A.C, Chevrier, B, Podjarny, A, Johnson, J, De Bear, J.S, Gough, G.R, Gilham, P.T, Moras, D. | | Deposit date: | 1990-02-01 | | Release date: | 1991-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic structure of an RNA helix: [U(UA)6A]2.
J.Mol.Biol., 209, 1989
|
|
8QVH
 
 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | 4-[(trans-4-{[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylcarbamoyl]amino}cyclohexyl)oxy]benzoic acid, Bifunctional epoxide hydrolase 2 | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 11, 2024
|
|
8QVF
 
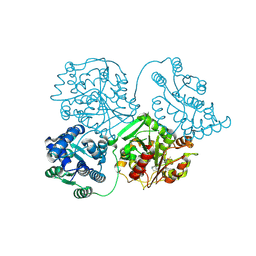 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | 1-(1-adamantyl)-3-(1-methylsulfonylpiperidin-4-yl)urea, Bifunctional epoxide hydrolase 2 | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 11, 2024
|
|
8QWG
 
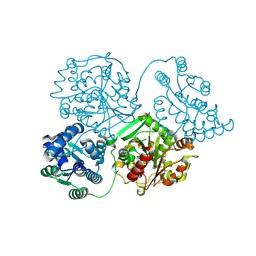 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | Bifunctional epoxide hydrolase 2, TRIETHYLENE GLYCOL | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 11, 2024
|
|
8QVL
 
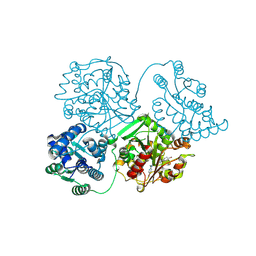 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | 2-[(5-BROMO-2-PYRIDYL)-METHYL-AMINO]ETHANOL, Bifunctional epoxide hydrolase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 11, 2024
|
|
8QVG
 
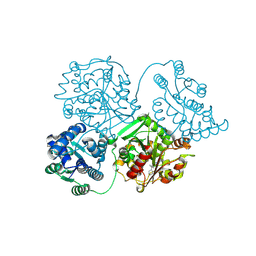 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | Bifunctional epoxide hydrolase 2, N-(3,3-DIPHENYLPROPYL)PYRROLIDINE-1-CARBOXAMIDE | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 11, 2024
|
|
8QVK
 
 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | Bifunctional epoxide hydrolase 2, N-(5,5-dioxodibenzothiophen-2-yl)-4,4-difluoro-piperidine-1-carboxamide | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 11, 2024
|
|
8QWI
 
 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | 2-(1H-BENZIMIDAZOL-2-YLSULFANYL)ETHANOL, Bifunctional epoxide hydrolase 2, SULFATE ION | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 11, 2024
|
|
8QVM
 
 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | Bifunctional epoxide hydrolase 2, TRIETHYLENE GLYCOL | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 11, 2024
|
|
8RNU
 
 | |
2XQ2
 
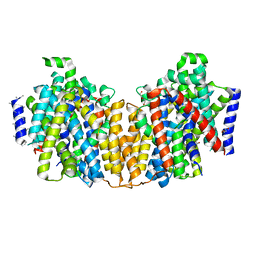 | | Structure of the K294A mutant of vSGLT | | Descriptor: | DI(HYDROXYETHYL)ETHER, SODIUM/GLUCOSE COTRANSPORTER | | Authors: | Watanabe, A, Choe, S, Chaptal, V, Rosenberg, J.M, Wright, E.M, Grabe, M, Abramson, J. | | Deposit date: | 2010-09-01 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Mechanism of Sodium and Substrate Release from the Binding Pocket of Vsglt
Nature, 468, 2010
|
|
2LW8
 
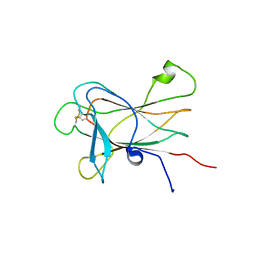 | |
4C69
 
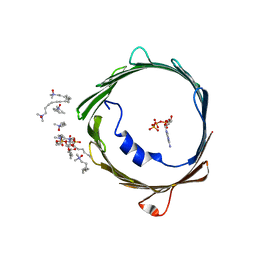 | | ATP binding to murine voltage-dependent anion channel 1 (mVDAC1). | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Paz, A, Colletier, J.P, Abramson, J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.277 Å) | | Cite: | Structure-Guided Simulations Illuminate the Mechanism of ATP Transport Through Vdac1.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2Y5Y
 
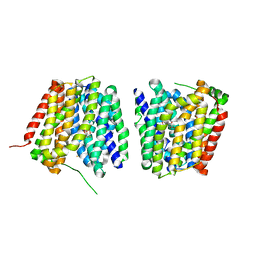 | | Crystal structure of LacY in complex with an affinity inactivator | | Descriptor: | 2-sulfanylethyl beta-D-galactopyranoside, BARIUM ION, LACTOSE PERMEASE | | Authors: | Chaptal, V, Kwon, S, Sawaya, M.R, Guan, L, Kaback, H.R, Abramson, J. | | Deposit date: | 2011-01-19 | | Release date: | 2011-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Crystal Structure of Lactose Permease in Complex with an Affinity Inactivator Yields Unique Insight Into Sugar Recognition.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
