7ZA2
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7Z3T
 
 | | Crystal structure of apo human Cathepsin L | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7ZAK
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
7ZFR
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide bound in the reverse direction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DP alpha chain (DPA1*02:01), MHC class II HLA-DP beta chain (DPB1*01:01), ... | | Authors: | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
7ZBB
 
 | | HaloTag with TRaQ-G-ctrl ligand | | Descriptor: | (E)-[7-azanyl-10-[2-carboxy-5-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]phenyl]-5,5-dimethyl-benzo[b][1]benzosilin-3-ylidene]-methyl-azanium, CHLORIDE ION, GLYCEROL, ... | | Authors: | Emmert, S, Rivera-Fuentes, P, Pojer, F, Lau, K. | | Deposit date: | 2022-03-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A locally activatable sensor for robust quantification of organellar glutathione.
Nat.Chem., 15, 2023
|
|
7ZBA
 
 | | HaloTag with Me-TRaQ-G ligand | | Descriptor: | 4-(7-azanyl-5,5-dimethyl-3-methylimino-benzo[b][1]benzosilin-10-yl)-N-[2-[2-(6-chloranylhexoxy)ethoxy]ethyl]-3-methyl-benzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Emmert, S, Rivera-Fuentes, P, Pojer, F, Lau, K. | | Deposit date: | 2022-03-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | A locally activatable sensor for robust quantification of organellar glutathione.
Nat.Chem., 15, 2023
|
|
7ZBD
 
 | | HaloTag with TRaQ-G ligand | | Descriptor: | (10R)-7-azanyl-N-[2-[2-(6-chloranylhexoxy)ethoxy]ethyl]-2'-cyano-5,5-dimethyl-3-(methylamino)-1'-oxidanylidene-spiro[benzo[b][1]benzosiline-10,3'-isoindole]-5'-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Emmert, S, Rivera-Fuentes, P, Pojer, F, Lau, K. | | Deposit date: | 2022-03-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A locally activatable sensor for robust quantification of organellar glutathione.
Nat.Chem., 15, 2023
|
|
8A67
 
 | |
8A1H
 
 | | Bacterial 6-4 photolyase from Vibrio cholerase | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-4 photolyase (FeS-BCP, ... | | Authors: | Essen, L.-O, Emmerich, H.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Functional Analysis of a Prokaryotic (6-4) Photolyase from the Aquatic Pathogen Vibrio Cholerae † .
Photochem.Photobiol., 99, 2023
|
|
8A7N
 
 | | Crystal Structure of human Brachyury G177D variant in complex with (S)-N-(3-aminopropyl)-3-((1-(2-fluorophenyl)-2-oxopyrrolidin-3-yl)amino)-N-methylbenzamide (CF-2-125) | | Descriptor: | N-(3-azanylpropyl)-3-[[(3S)-1-(2-fluorophenyl)-2-oxidanylidene-pyrrolidin-3-yl]amino]-N-methyl-benzamide, PHOSPHATE ION, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-06-21 | | Release date: | 2022-10-05 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into human brachyury DNA recognition and discovery of progressible binders for cancer therapy.
Nat Commun, 16, 2025
|
|
8AHW
 
 | | Structure of DCS-resistant variant D322N of alanine racemase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Alanine racemase, GLYCEROL | | Authors: | de Chiara, C, Prosser, G, Ogrodowicz, R.W, de Carvalho, L.P.S. | | Deposit date: | 2022-07-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of the d-Cycloserine-Resistant Variant D322N of Alanine Racemase from Mycobacterium tuberculosis .
Acs Bio Med Chem Au, 3, 2023
|
|
8S09
 
 | | H. sapiens MCM2-7 double hexamer bound to double stranded DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (45-mer), ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | MCM double hexamer loading visualized with human proteins.
Nature, 636, 2024
|
|
8S0C
 
 | | H. sapiens ORC1-5 bound to double stranded DNA as part of the MCM-ORC complex | | Descriptor: | DNA (26-mer), Isoform 2 of Origin recognition complex subunit 3, MAGNESIUM ION, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | MCM double hexamer loading visualized with human proteins.
Nature, 636, 2024
|
|
8S0B
 
 | | H. sapiens MCM bound to double stranded DNA and ORC6 as part of the MCM-ORC complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (45-mer), DNA replication licensing factor MCM2, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | MCM double hexamer loading visualized with human proteins.
Nature, 636, 2024
|
|
8S0D
 
 | | H. sapiens MCM bound to double stranded DNA and ORC1-6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (58-mer), DNA replication licensing factor MCM2, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | MCM double hexamer loading visualized with human proteins.
Nature, 636, 2024
|
|
8S0F
 
 | | H. sapiens OC1M bound to double stranded DNA | | Descriptor: | DNA (39-mer), DNA replication factor Cdt1, DNA replication licensing factor MCM2, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | MCM double hexamer loading visualized with human proteins.
Nature, 636, 2024
|
|
8S0E
 
 | | H. sapiens OCCM bound to double stranded DNA | | Descriptor: | Cell division control protein 6 homolog, DNA (39-mer), DNA replication factor Cdt1, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | MCM double hexamer loading visualized with human proteins.
Nature, 636, 2024
|
|
8S0A
 
 | | H. sapiens MCM2-7 hexamer bound to double stranded DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (22-mer), ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | MCM double hexamer loading visualized with human proteins.
Nature, 636, 2024
|
|
8RLY
 
 | | E. coli endonuclease IV complexed with sulfate, catalytic Fe2+ | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Endonuclease 4, ... | | Authors: | Saper, M.A, Paterson, N.G, Kirillov, S, Rouvinski, A. | | Deposit date: | 2024-01-04 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Octahedral Iron in Catalytic Sites of Endonuclease IV from Staphylococcus aureus and Escherichia coli .
Biochemistry, 64, 2025
|
|
8RZU
 
 | | Structure of human SETD2 L1609P mutant in complex with SAM and H3K36M peptide | | Descriptor: | Histone H3, Histone-lysine N-methyltransferase SETD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Mechaly, A.E, Michail, C, Haouz, A, Rodrigues-Lima, F. | | Deposit date: | 2024-02-13 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of human SETD2 L1609P mutant in complex with SAM and H3K36M peptide
To Be Published
|
|
8SP7
 
 | |
8SBH
 
 | | YeiE effector binding domain from E. coli | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Momany, C, Nune, M, Brondani, J.C, Afful, D, Neidle, E. | | Deposit date: | 2023-04-03 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | FinR, a LysR-type transcriptional regulator involved in sulfur homeostasis with homologs in diverse microorganisms
To Be Published
|
|
8S8A
 
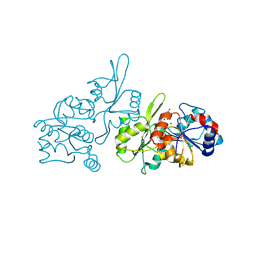 | | Human pyridoxal phosphatase in complex with 7,8-dihydroxyflavone without phosphate | | Descriptor: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, CHLORIDE ION, Chronophin, ... | | Authors: | Brenner, M, Gohla, A, Schindelin, H. | | Deposit date: | 2024-03-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
8TJG
 
 | |
8TUA
 
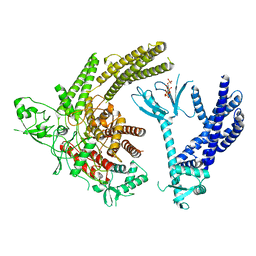 | | Full-length P-Rex1 in complex with inositol 1,3,4,5-tetrakisphosphate (IP4) | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2023-08-15 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Full-length P-Rex1 in complex with inositol 1,3,4,5-tetrakisphosphate (IP4)
Elife, 2024
|
|
