6X2J
 
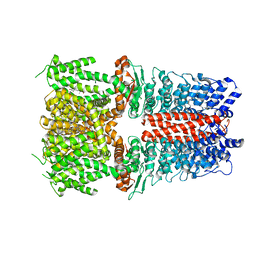 | | Structure of human TRPA1 in complex with agonist GNE551 | | Descriptor: | 5-amino-1-[(4-bromo-2-fluorophenyl)methyl]-N-(2,5-dimethoxyphenyl)-1H-1,2,3-triazole-4-carboxamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Chen, H. | | Deposit date: | 2020-05-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A Non-covalent Ligand Reveals Biased Agonism of the TRPA1 Ion Channel.
Neuron, 109, 2021
|
|
7XFA
 
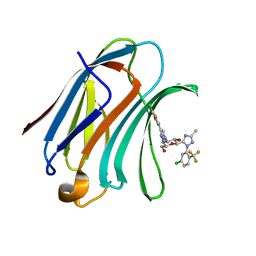 | | Structure of human Galectin-3 CRD in complex with monosaccharide inhibitor | | Descriptor: | (2~{S},3~{R},4~{R},5~{R},6~{R})-4-[4-[4-chloranyl-3,5-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-[2-[5-chloranyl-2-(trifluoromethyl)phenyl]-5-methyl-1,2,4-triazol-3-yl]-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | Authors: | Shukla, J, Raman, S, Ghosh, K. | | Deposit date: | 2022-04-01 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Identification of Monosaccharide Derivatives as Potent, Selective, and Orally Bioavailable Inhibitors of Human and Mouse Galectin-3.
J.Med.Chem., 65, 2022
|
|
8FK2
 
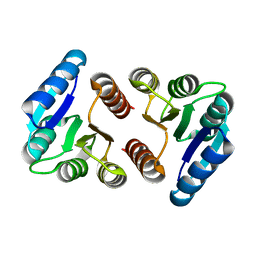 | | The N-terminal VicR from Streptococcus mutans | | Descriptor: | Putative response regulator CovR VicR-like protein | | Authors: | Zhang, H, Wu, H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Small Molecule Attenuates Bacterial Virulence by Targeting Conserved Response Regulator.
Mbio, 14, 2023
|
|
7K7Q
 
 | |
7K7O
 
 | |
6ILW
 
 | | Crystal structure of PETase from Ideonella sakaiensis | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SODIUM ION | | Authors: | Liu, C.C, Shi, C. | | Deposit date: | 2018-10-19 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.575 Å) | | Cite: | Structural and functional characterization of polyethylene terephthalate hydrolase from Ideonella sakaiensis.
Biochem. Biophys. Res. Commun., 508, 2019
|
|
3MVL
 
 | | P38 Alpha Map Kinase complexed with pyrrolotriazine inhibitor 7K | | Descriptor: | 4-{[5-(cyclopropylcarbamoyl)-2-methylphenyl]amino}-5-methyl-N-propylpyrrolo[2,1-f][1,2,4]triazine-6-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2010-05-04 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 4-(5-(Cyclopropylcarbamoyl)-2-Methylphenylamino)-5-Methyl-Npropylpyrrolo[1,2-F][1,2,4] Triazine-6-Carboxamide (Bms-582949), a Clinical P38 Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
J.Med.Chem., 53, 2010
|
|
6ILX
 
 | | Crystal structure of PETase W159F mutant from Ideonella sakaiensis | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SODIUM ION | | Authors: | Liu, C.C, Shi, C. | | Deposit date: | 2018-10-20 | | Release date: | 2019-03-27 | | Last modified: | 2020-09-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional characterization of polyethylene terephthalate hydrolase from Ideonella sakaiensis.
Biochem. Biophys. Res. Commun., 508, 2019
|
|
3MVM
 
 | | P38 Alpha Map Kinase complexed with pyrrolotriazine inhibitor 7V | | Descriptor: | 4-{[5-(isoxazol-3-ylcarbamoyl)-2-methylphenyl]amino}-5-methyl-N-propylpyrrolo[2,1-f][1,2,4]triazine-6-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2010-05-04 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-(5-(Cyclopropylcarbamoyl)-2-Methylphenylamino)-5-Methyl-Npropylpyrrolo[1,2-F][1,2,4] Triazine-6-Carboxamide (Bms-582949), a Clinical P38? Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
J.Med.Chem., 53, 2010
|
|
7EQL
 
 | | Crystal structure of (+)-pulegone reductase from Mentha piperita | | Descriptor: | (+)-pulegone reductase | | Authors: | Lin, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Functional Characterization and Structural Insights Into Stereoselectivity of Pulegone Reductase in Menthol Biosynthesis.
Front Plant Sci, 12, 2021
|
|
5BNQ
 
 | | Crystal structure of hRANKL-mRANK complex | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Ren, J. | | Deposit date: | 2015-05-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A RANKL mutant used as an inter-species vaccine for efficient immunotherapy of osteoporosis.
Sci Rep, 5, 2015
|
|
8ILU
 
 | | Crystal structure of mouse Galectin-3 in complex with small molecule inhibitor | | Descriptor: | (2R,3R,4R,5R,6S)-2-(hydroxymethyl)-6-[2-(2-methyl-1,3-benzothiazol-6-yl)-1,2,4-triazol-3-yl]-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, SODIUM ION, ... | | Authors: | Kumar, A, Jinal, S, Raman, S, Ghosh, K. | | Deposit date: | 2023-03-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of benzothiazole derived monosaccharides as potent, selective, and orally bioavailable inhibitors of human and mouse galectin-3; a rare example of using a S···O binding interaction for drug design.
Bioorg.Med.Chem., 101, 2024
|
|
6JP1
 
 | |
6J9R
 
 | |
6JYZ
 
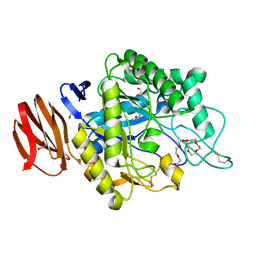 | | Crystal structure of endogalactoceramidase | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liuqing, C, Yan, F. | | Deposit date: | 2019-04-29 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of an endogalactosylceramidase from Rhodococcus hoagii 103S reveals the molecular basis of its substrate specificity.
J.Struct.Biol., 208, 2019
|
|
7QNY
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with COVOX-58 and COVOX-158 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7QNW
 
 | | The receptor binding domain of SARS-CoV-2 Omicron variant spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Beta-55 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7QNX
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-55 heavy chain, Beta-55 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7LAW
 
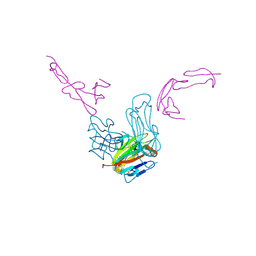 | | crystal structure of GITR complex with GITR-L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, Tumor necrosis factor receptor superfamily member 18 | | Authors: | Longenecker, K.L, Rogers, B, Bigelow, L, Judge, R.A, Alvarez, H. | | Deposit date: | 2021-01-07 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | An anti-PD-1-GITR-L bispecific agonist induces GITR clustering-mediated T cell activation for cancer immunotherapy.
Nat Cancer, 3, 2022
|
|
8C3V
 
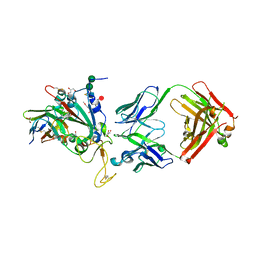 | | SARS-CoV-2 Delta-RBD complexed with BA.2-13 Fab and C1 nanobody | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, BA.2-13 heavy chain, BA.2-13 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-12-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
8BBN
 
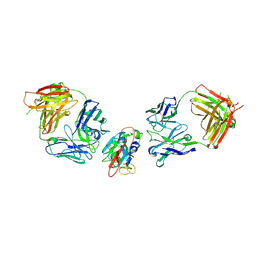 | | SARS-CoV-2 Delta-RBD complexed with BA.2-10 and EY6A Fabs | | Descriptor: | BA.2-10 heavy chain, BA.2-10 light chain, EY6A Heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
8BBO
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.2-36 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IGH@ protein, Immunoglobulin kappa light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
8BCZ
 
 | | SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45 | | Descriptor: | BA.2-23 heavy chain, BA.2-23 light chain, BA.2-36 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-17 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
6HSR
 
 | |
5J8V
 
 | |
