7AQJ
 
 | | Structure of SARS-CoV-2 Main Protease bound to Triglycidyl isocyanurate. | | Descriptor: | 1-[(2~{R})-2-oxidanylpropyl]-3-[[(2~{R})-oxiran-2-yl]methyl]-5-[[(2~{S})-oxiran-2-yl]methyl]-1,3,5-triazinane-2,4,6-trione, 3C-like proteinase, Triglycidyl isocyanurate | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AOL
 
 | | Structure of SARS-CoV-2 Main Protease bound to Climbazole | | Descriptor: | (1~{S})-1-(4-chloranylphenoxy)-1-imidazol-1-yl-3,3-dimethyl-butan-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7YXU
 
 | | Crystal structure of agonistic antibody 1618 fab domain bound to human 4-1BB. | | Descriptor: | MANGANESE (II) ION, Tumor necrosis factor receptor superfamily member 9, heavy chain of Fab, ... | | Authors: | Hakansson, M, Rose, N, Petersson, J, Enell Smith, K, Thorolfsson, M, von Schantz, L. | | Deposit date: | 2022-02-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Bispecific Tumor Antigen-Conditional 4-1BB x 5T4 Agonist, ALG.APV-527, Mediates Strong T-Cell Activation and Potent Antitumor Activity in Preclinical Studies.
Mol.Cancer Ther., 22, 2023
|
|
7D5Z
 
 | | Crystal structure of EBV gH/gL bound with neutralizing antibody 1D8 | | Descriptor: | Envelope glycoprotein H, Envelope glycoprotein L, heavy chain of 1D8, ... | | Authors: | Zhu, Q, Shan, S, Yu, J, Wang, X, Zhang, L, Zeng, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | A Neutralizing Antibody Targeting a New Site of Vulnerability on Epstein-Barr Virus gH/gL Protects against Dual-Tropic Infection
To Be Published
|
|
8IF8
 
 | | Arabinosyltransferase AftA | | Descriptor: | CALCIUM ION, Galactan 5-O-arabinofuranosyltransferase | | Authors: | Gong, Y.C, Rao, Z.H, Zhang, L. | | Deposit date: | 2023-02-17 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the priming arabinosyltransferase AftA required for AG biosynthesis of Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7B83
 
 | | Structure of SARS-CoV-2 Main Protease bound to pyrithione zinc | | Descriptor: | 3C-like proteinase, 9-oxa-7-thia-1-azonia-8$l^{2}-zincabicyclo[4.3.0]nona-1,3,5-triene, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8H4Q
 
 | |
8H4H
 
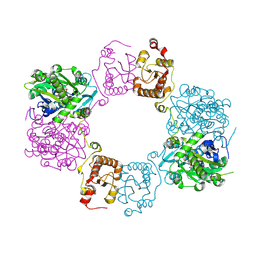 | |
2RJG
 
 | | Crystal structure of biosynthetic alaine racemase from Escherichia coli | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-10-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
2RJH
 
 | | Crystal structure of biosynthetic alaine racemase in D-cycloserine-bound form from Escherichia coli | | Descriptor: | Alanine racemase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-10-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
7CUQ
 
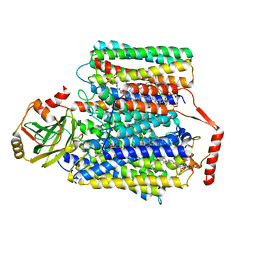 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-24 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUB
 
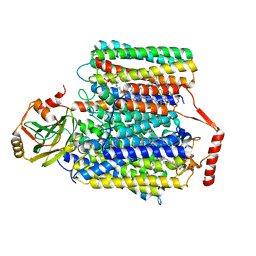 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-22 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUW
 
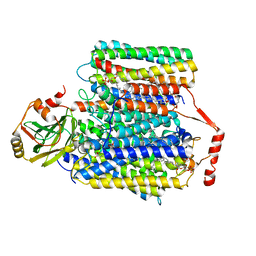 | | Ubiquinol Binding Site of Cytochrome bo3 from Escherichia coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-25 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4ZA1
 
 | | Crystal Structure of NosA Involved in Nosiheptide Biosynthesis | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NosA | | Authors: | Liu, S, Guo, H, Zhang, T, Han, L, Yao, P, Zhang, Y, Rong, N, Yu, Y, Lan, W, Wang, C, Ding, J, Wang, R, Liu, W, Cao, C. | | Deposit date: | 2015-04-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based Mechanistic Insights into Terminal Amide Synthase in Nosiheptide-Represented Thiopeptides Biosynthesis
Sci Rep, 5, 2015
|
|
5BOF
 
 | | Crystal Structure of Staphylococcus aureus Enolase | | Descriptor: | Enolase, MAGNESIUM ION, SULFATE ION | | Authors: | Wu, Y.F, Wang, C.L, Wu, M.H, Han, L, Zhang, X, Zang, J.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5BOE
 
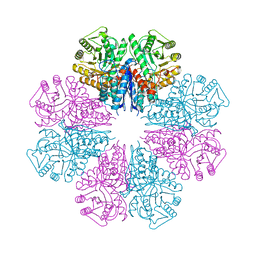 | | Crystal structure of Staphylococcus aureus enolase in complex with PEP | | Descriptor: | Enolase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wang, C.L, Wu, Y.F, Han, L, Wu, M.H, Zhang, X, Zang, J.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2FXZ
 
 | |
2FXY
 
 | |
5XJX
 
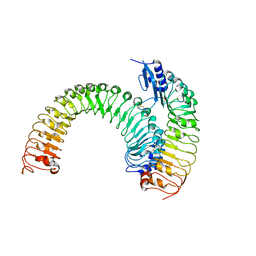 | | Pre-formed plant receptor ERL1-TMM complex | | Descriptor: | LRR receptor-like serine/threonine-protein kinase ERL1, Protein TOO MANY MOUTHS | | Authors: | Chai, J, Lin, G, Zhang, L, Han, Z, Yang, X, Liu, W, Qi, Y, Chang, J, Li, E. | | Deposit date: | 2017-05-04 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.055 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
5XJO
 
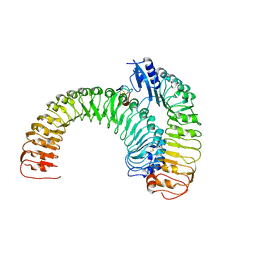 | | Plant receptor ERL1-TMM in complex with peptide EPF1 | | Descriptor: | LRR receptor-like serine/threonine-protein kinase ERL1, Protein EPIDERMAL PATTERNING FACTOR 1, Protein TOO MANY MOUTHS | | Authors: | Chai, J, Lin, G, Zhang, L, Han, Z, Shpak, E.D, Yang, X. | | Deposit date: | 2017-05-03 | | Release date: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.626 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
8JI8
 
 | | Crystal Structure of Prophenoloxidase PPO6 chimeric mutant (F215EASNRAIVD224 to G215DGPDSVVR223) from Aedes aegypti | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, ... | | Authors: | Zhu, X, Zhang, L, Yang, X, Bao, P, Ren, D, Han, Q. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mosquitoes have evolved two types of prophenoloxidases
To Be Published
|
|
8JIB
 
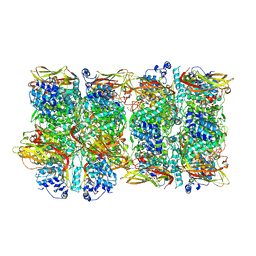 | | Crystal Structure of Prophenoloxidase PPO6 from Aedes aegypti | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, TK receptor | | Authors: | Zhu, X, Zhang, L, Yang, X, Bao, P, Ren, D, Han, Q. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Mosquitoes have evolved two types of prophenoloxidases
To Be Published
|
|
6M54
 
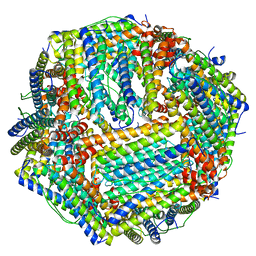 | | Human apo ferritin frozen on TEM grid with Amorphous nickel titanium alloy supporting film | | Descriptor: | FE (II) ION, Ferritin heavy chain | | Authors: | Huang, X, Zhang, L, Wen, Z, Chen, H, Li, S, Ji, G, Yin, C, Sun, F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation.
Prog.Biophys.Mol.Biol., 156, 2020
|
|
5B5K
 
 | | Crystal structure of Izumo1, the mammalian sperm ligand for egg Juno | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Izumo sperm-egg fusion protein 1 | | Authors: | Nishimura, K, Han, L, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-05-11 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of sperm Izumo1 reveals unexpected similarities with Plasmodium invasion proteins.
Curr.Biol., 26, 2016
|
|
6WYF
 
 | |
