8TT4
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=6.0 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TT5
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=8.3 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TSU
 
 | |
8TSX
 
 | |
8TT1
 
 | |
8TWI
 
 | | Cryo-EM structure of the PP2A:B55-FAM122A complex, PP2Ac body | | Descriptor: | FE (III) ION, PPP2R1A-PPP2R2A-interacting phosphatase regulator 1, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Fuller, J.R, Padi, S.K.R, Peti, W, Page, R. | | Deposit date: | 2023-08-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Nature, 625, 2024
|
|
8TWE
 
 | | Cryo-EM structure of the PP2A:B55-FAM122A complex, B55 body | | Descriptor: | PPP2R1A-PPP2R2A-interacting phosphatase regulator 1, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Fuller, J.R, Padi, S.K.R, Peti, W, Page, R. | | Deposit date: | 2023-08-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Nature, 625, 2024
|
|
8TBW
 
 | | Human Class I MHC HLA-A2 in complex with sorting nexin 24 (127-135) peptide KLSHQPVLL | | Descriptor: | Beta-2-microglobulin, HLA-A*02:01 alpha chain, NITRATE ION, ... | | Authors: | Arbuiso, A, Weiss, L.I, Brambley, C.A, Ma, J, Keller, G.L.J, Ayres, C.M, Baker, B.M. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Accurate modeling of peptide-MHC structures with AlphaFold.
Structure, 32, 2024
|
|
8TBV
 
 | | Human Class I MHC HLA-A2 in complex with sorting nexin 24 (127-135) neoantigen KLSHQLVLL | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Arbuiso, A, Weiss, L.I, Brambley, C.A, Ma, J, Keller, G.L.J, Ayres, C.M, Baker, B.M. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Accurate modeling of peptide-MHC structures with AlphaFold.
Structure, 32, 2024
|
|
8TUA
 
 | | Full-length P-Rex1 in complex with inositol 1,3,4,5-tetrakisphosphate (IP4) | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2023-08-15 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Full-length P-Rex1 in complex with inositol 1,3,4,5-tetrakisphosphate (IP4)
Elife, 2024
|
|
8U9G
 
 | | Human Class I MHC HLA-A2 bound to sorting nexin 24 (127-135) neoantigen KLSHQLVLL | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Arbuiso, A, Weiss, L.I, Brambley, C.A, Ma, J, Keller, G.L.J, Ayres, C.M, Baker, B.M. | | Deposit date: | 2023-09-19 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Accurate modeling of peptide-MHC structures with AlphaFold.
Structure, 32, 2024
|
|
8TSY
 
 | |
8TT2
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=5.4 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TSZ
 
 | |
8V1J
 
 | |
8F43
 
 | | HNH Nuclease Domain from G. stearothermophilus Cas9, K597A mutant | | Descriptor: | CRISPR-associated endonuclease Cas9 | | Authors: | D'Ordine, A.M, Belato, H.B, Lisi, G.P, Jogl, G. | | Deposit date: | 2022-11-10 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Disruption of electrostatic contacts in the HNH nuclease from a thermophilic Cas9 rewires allosteric motions and enhances high-temperature DNA cleavage.
J.Chem.Phys., 157, 2022
|
|
8FOI
 
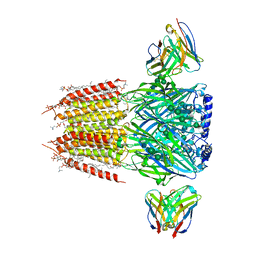 | | Native GABA-A receptor from the mouse brain, alpha1-beta2-gamma2 subtype, in complex with GABA and allopregnanolone | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, DODECANE, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2022-12-30 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8F6U
 
 | |
8F6V
 
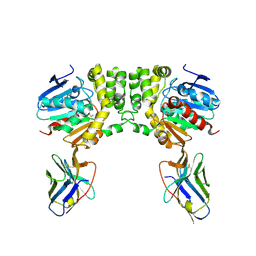 | |
8G6C
 
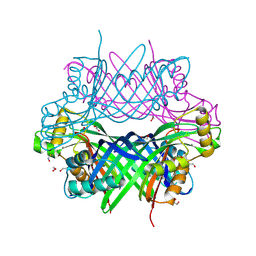 | |
8GAR
 
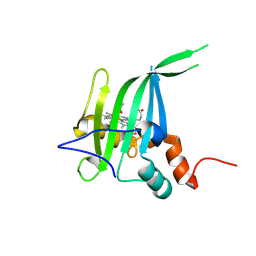 | |
8G5G
 
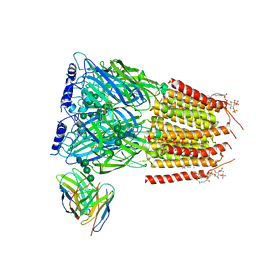 | | Native GABA-A receptor from the mouse brain, meta-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA, Zolpidem, and endogenous neurosteroid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-13 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G4X
 
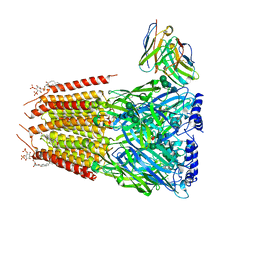 | | Native GABA-A receptor from the mouse brain, meta-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA and allopregnanolone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G5F
 
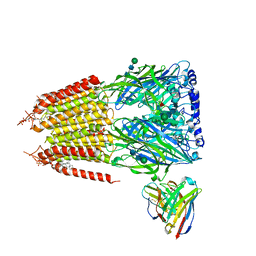 | | Native GABA-A receptor from the mouse brain, ortho-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA and allopregnanolone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-13 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G4O
 
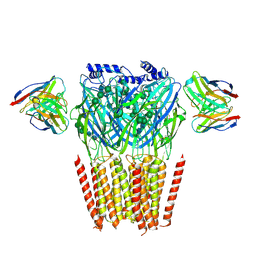 | | Native GABA-A receptor from the mouse brain, alpha1-beta2-gamma2 subtype, in complex with didesethylflurazepam and endogenous GABA | | Descriptor: | (5M)-1-(2-aminoethyl)-7-chloro-5-(2-fluorophenyl)-1,3-dihydro-2H-1,4-benzodiazepin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
