8ZHL
 
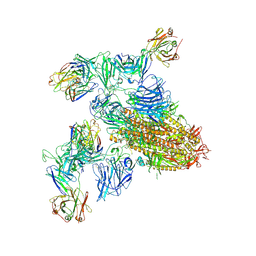 | | SARS-CoV-2 spike trimer (6P) in complex with two H18 and two R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein helping to drive the genesis of Omicron variants
To Be Published
|
|
5VLZ
 
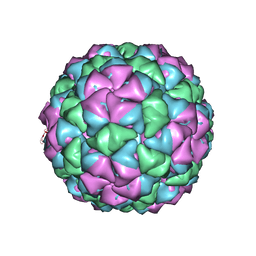
 | | Backbone model for phage Qbeta capsid | | Descriptor: | Capsid protein, Maturation protein A2 | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of Q beta virions, virus-like particles, and the Q beta-MurA complex reveal internal coat proteins and the mechanism of host lysis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KIP
 
 | |
4WCW
 
 | | Ribosomal silencing factor during starvation or stationary phase (RsfS) from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, Ribosomal silencing factor RsfS | | Authors: | Li, X, Sun, Q, Jiang, C, Yang, K, Hung, L, Zhang, J, Sacchettini, J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Ribosomal Silencing Factor Bound to Mycobacterium tuberculosis Ribosome.
Structure, 23, 2015
|
|
4F5Y
 
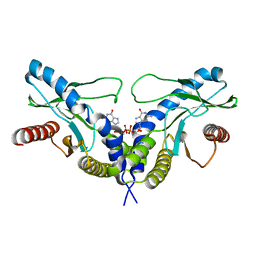 | | Crystal structure of human STING CTD complex with C-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Transmembrane protein 173 | | Authors: | Gu, L, Shang, G, Zhu, D, Li, N, Zhang, J, Zhu, C, Lu, D, Liu, C, Yu, Q, Zhao, Y, Xu, S. | | Deposit date: | 2012-05-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP
Nat.Struct.Mol.Biol., 19, 2012
|
|
4F5W
 
 | | Crystal structure of ligand free human STING CTD | | Descriptor: | CALCIUM ION, Transmembrane protein 173 | | Authors: | Gu, L, Shang, G, Zhu, D, Li, N, Zhang, J, Zhu, C, Lu, D, Liu, C, Yu, Q, Zhao, Y, Xu, S. | | Deposit date: | 2012-05-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP
Nat.Struct.Mol.Biol., 19, 2012
|
|
5B6G
 
 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, GLYCEROL, PHQ-ALA-GLY-GLU-ALA-XYC-TYR-GLU, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
8Y2T
 
 | | Crystal structure of 3C protease from coxsackievirus B3 | | Descriptor: | Protease 3C | | Authors: | Jiang, H.H, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the 3C proteases from Coxsackievirus B3 and B4.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8Y2U
 
 | | Crystal structure of 3C protease from coxsackievirus B4 | | Descriptor: | Protease 3C | | Authors: | Jiang, H.H, Lin, C, Zhang, J, Li, J. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of the 3C proteases from Coxsackievirus B3 and B4.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
9C9Q
 
 | |
9C9R
 
 | |
5IZ8
 
 | | Protein-protein interaction | | Descriptor: | ACE-ALA-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein, TRIETHYLENE GLYCOL | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZA
 
 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-TRP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZ9
 
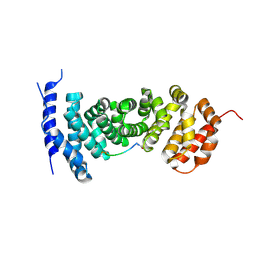 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZ6
 
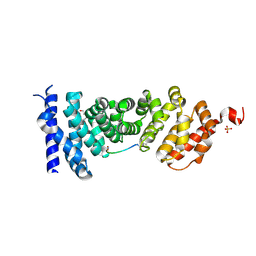 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, DI(HYDROXYETHYL)ETHER, PHQ-ALA-GLY-GLU-ALA-LEU-TYR-GLU-NH2, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
8WZQ
 
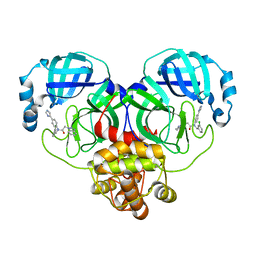 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with CCF0058981 | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(1~{H}-imidazol-5-yl)phenyl]ethanamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Jiang, H.H, Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Biochem.Biophys.Res.Commun., 692, 2024
|
|
1YOP
 
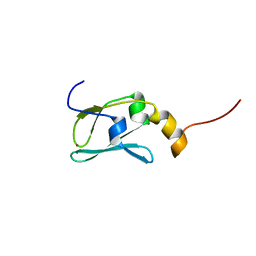 | | The solution structure of Kti11p | | Descriptor: | Kti11p, ZINC ION | | Authors: | Sun, J, Zhang, J, Wu, F, Xu, C, Li, S, Zhao, W, Wu, Z, Wu, J, Zhou, C.-Z, Shi, Y. | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Kti11p from Saccharomyces cerevisiae reveals a novel zinc-binding module.
Biochemistry, 44, 2005
|
|
2MOX
 
 | | solution structure of tandem SH3 domain of Sorbin and SH3 domain-containing protein 1 | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Zhao, D, Wang, C, Zhang, J, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2014-05-07 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
4EQ8
 
 | | Crystal structure of PA1844 from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, Putative uncharacterized protein | | Authors: | Shang, G, Li, N, Zhang, J, Lu, D, Yu, Q, Zhao, Y, Liu, X, Xu, S, Gu, L. | | Deposit date: | 2012-04-18 | | Release date: | 2012-09-12 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structural insight into how Pseudomonas aeruginosa peptidoglycanhydrolase Tse1 and its immunity protein Tsi1 function.
Biochem.J., 448, 2012
|
|
4EQA
 
 | | Crystal structure of PA1844 in complex with PA1845 from Pseudomonas aeruginosa PAO1 | | Descriptor: | Putative uncharacterized protein | | Authors: | Shang, G, Li, N, Zhang, J, Lu, D, Yu, Q, Zhao, Y, Liu, X, Xu, S, Gu, L. | | Deposit date: | 2012-04-18 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insight into how Pseudomonas aeruginosa peptidoglycanhydrolase Tse1 and its immunity protein Tsi1 function.
Biochem.J., 448, 2012
|
|
8VU0
 
 | |
2L89
 
 | |
8ZQ3
 
 | | Structure of ORP1L-RAB7A in the presence of GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Oxysterol-binding protein-related protein 1, Ras-related protein Rab-7a | | Authors: | Lu, Q, Zhang, J, Xu, C. | | Deposit date: | 2024-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of GDP-bound Rab7 Q67L in complex with ORP1L.
Biochem.Biophys.Res.Commun., 725, 2024
|
|
8EPE
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*CP*CP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*G)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPB
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*GP*GP*TP*GP*GP*TP*TP*CP*GP*A)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*GP*CP*CP*GP*AP*AP*CP*CP*T)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
