7X73
 
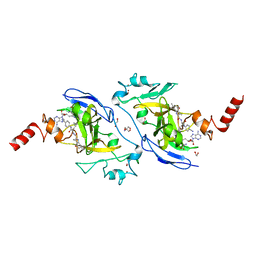 | | Structure of G9a in complex with RK-701 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-03-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A specific G9a inhibitor unveils BGLT3 lncRNA as a universal mediator of chemically induced fetal globin gene expression.
Nat Commun, 14, 2023
|
|
2DO6
 
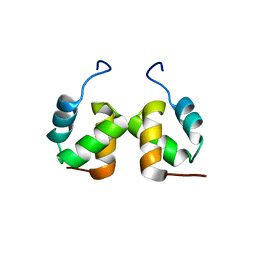 | | Solution structure of RSGI RUH-065, a UBA domain from human cDNA | | Descriptor: | E3 ubiquitin-protein ligase CBL-B | | Authors: | Hamada, T, Hirota, H, Lin, Y.-J, Guntert, P, Sato, M, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-27 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RSGI RUH-065, a UBA domain from human cDNA
To be Published
|
|
9KLB
 
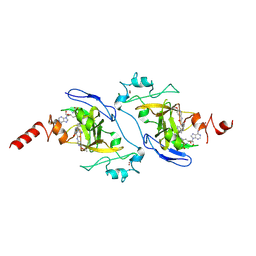 | | G9a in complex with RK-133232 (compound 16g) | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, SINEFUNGIN, ZINC ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Ihara, K, Shirouzu, M, Umehara, T. | | Deposit date: | 2024-11-14 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of potent substrate-type lysine methyltransferase G9a inhibitors for the treatment of sickle cell disease.
Eur.J.Med.Chem., 293, 2025
|
|
9KLC
 
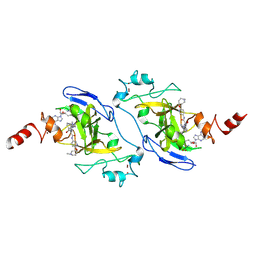 | | G9a in complex with the S-isomer of RK-131902 (racemic compound rac-10a) | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, SINEFUNGIN, ZINC ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Ihara, K, Shirouzu, M, Umehara, T. | | Deposit date: | 2024-11-14 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of potent substrate-type lysine methyltransferase G9a inhibitors for the treatment of sickle cell disease.
Eur.J.Med.Chem., 293, 2025
|
|
5AVL
 
 | | Crystal structure of LXRalpha in complex with tert-butyl benzoate analog, compound 32b | | Descriptor: | 2-[4-[4-[[2-[(2-methylpropan-2-yl)oxycarbonyl]-3-oxidanyl-4-(trifluoromethyl)phenyl]methoxy]phenyl]phenyl]ethanoic acid, Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha | | Authors: | Matsui, Y, Hanzawa, H, Tamaki, K. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and structure-guided optimization of tert-butyl 6-(phenoxymethyl)-3-(trifluoromethyl)benzoates as liver X receptor agonists
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5AVI
 
 | | Crystal structure of LXRalpha in complex with tert-butyl benzoate analog, compound 4 | | Descriptor: | Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, tert-butyl 2-[[4-[ethanoyl(methyl)amino]phenoxy]methyl]-5-(trifluoromethyl)benzoate | | Authors: | Matsui, Y, Hanzawa, H, Tamaki, K. | | Deposit date: | 2015-06-16 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and structure-guided optimization of tert-butyl 6-(phenoxymethyl)-3-(trifluoromethyl)benzoates as liver X receptor agonists
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1E79
 
 | | Bovine F1-ATPase inhibited by DCCD (dicyclohexylcarbodiimide) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP SYNTHASE ALPHA CHAIN HEART ISOFORM, ... | | Authors: | Gibbons, C, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2000-08-25 | | Release date: | 2000-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of the Central Stalk in Bovine F(1)-ATPase at 2.4 A Resolution.
Nat.Struct.Biol., 7, 2000
|
|
2DVX
 
 | |
2DVT
 
 | | Crystal Structure of 2,6-Dihydroxybenzoate Decarboxylase from Rhizobium | | Descriptor: | Thermophilic reversible gamma-resorcylate decarboxylase, ZINC ION | | Authors: | Goto, M. | | Deposit date: | 2006-08-01 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Nonoxidative Zn-Dependent 2,6-Dihydroxybenzoate (gamma-Resorcylate) Decarboxylase
from Rhizobium sp. Strain MTP-10005
To be Published
|
|
7CP0
 
 | | Crystal Structure of double mutant Y115E Y117E human Secretory Glutaminyl Cyclase | | Descriptor: | 1,2-ETHANEDIOL, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Dileep, K.V, Ihara, K, Sakai, N, Shirozu, M, Zhang, K.Y.J. | | Deposit date: | 2020-08-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Piperidine-4-carboxamide as a new scaffold for designing secretory glutaminyl cyclase inhibitors.
Int.J.Biol.Macromol., 170, 2020
|
|
7COZ
 
 | | Crystal Structure of double mutant Y115E Y117E human Secretory Glutaminyl Cyclase in complex with LSB-41 | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-(2-methyl-4-thiophen-2-yl-1,3-thiazol-5-yl)propanoyl]piperidine-4-carboxamide, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Dileep, K.V, Ihara, K, Sakai, N, Shirozu, M, Zhang, K.Y.J. | | Deposit date: | 2020-08-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Piperidine-4-carboxamide as a new scaffold for designing secretory glutaminyl cyclase inhibitors.
Int.J.Biol.Macromol., 170, 2020
|
|
1IVZ
 
 | | Solution structure of the SEA domain from murine hypothetical protein homologous to human mucin 16 | | Descriptor: | hypothetical protein 1110008I14RIK | | Authors: | Maeda, T, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-04-02 | | Release date: | 2002-10-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SEA domain from the murine homologue of ovarian cancer antigen CA125 (MUC16)
J.Biol.Chem., 279, 2004
|
|
2DVU
 
 | |
7BOT
 
 | | Human SIRT2 in complex with myristoyl thiourea inhibitor, No.23 | | Descriptor: | N-dodecylmethanethioamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | Authors: | Kudo, N, Olsen, C.A, Minoru, Y. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism-based inhibitors of SIRT2: structure-activity relationship, X-ray structures, target engagement, regulation of alpha-tubulin acetylation and inhibition of breast cancer cell migration.
Rsc Chem Biol, 2, 2021
|
|
7BOS
 
 | | Human SIRT2 in complex with myristoyl thiourea inhibitor, No.13 | | Descriptor: | Myristoyl thiourea inhibitor, No.13, N-dodecylmethanethioamide, ... | | Authors: | Kudo, N, Olsen, C.A, Minoru, Y. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism-based inhibitors of SIRT2: structure-activity relationship, X-ray structures, target engagement, regulation of alpha-tubulin acetylation and inhibition of breast cancer cell migration.
Rsc Chem Biol, 2, 2021
|
|
1BHI
 
 | | STRUCTURE OF TRANSACTIVATION DOMAIN OF CRE-BP1/ATF-2, NMR, 20 STRUCTURES | | Descriptor: | CRE-BP1 | | Authors: | Nagadoi, A, Nakazawa, K, Uda, H, Maekawa, T, Ishii, S, Nishimura, Y. | | Deposit date: | 1998-06-09 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the transactivation domain of ATF-2 comprising a zinc finger-like subdomain and a flexible subdomain.
J.Mol.Biol., 287, 1999
|
|
1UKX
 
 | | Solution structure of the RWD domain of mouse GCN2 | | Descriptor: | GCN2 eIF2alpha kinase | | Authors: | Nameki, N, Yoneyama, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RWD domain of the mouse GCN2 protein.
Protein Sci., 13, 2004
|
|
9K7O
 
 | | Coprinopsis cinerea GH131 protein CcGH131B | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glycoside hydrolase 131 catalytic N-terminal domain-containing protein | | Authors: | Shiojima, Y, Tonozuka, T. | | Deposit date: | 2024-10-24 | | Release date: | 2024-11-06 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of CcGH131B, a Protein Belonging to Glycoside Hydrolase Family 131 from the Basidiomycete Coprinopsis cinerea .
J Appl Glycosci (1999), 72, 2025
|
|
9K7M
 
 | | Coprinopsis cinerea GH131 protein CcGH131B E161A in complex with cellobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Glycoside hydrolase 131 catalytic N-terminal domain-containing protein, ... | | Authors: | Shiojima, Y, Tonozuka, T. | | Deposit date: | 2024-10-24 | | Release date: | 2024-11-06 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of CcGH131B, a Protein Belonging to Glycoside Hydrolase Family 131 from the Basidiomycete Coprinopsis cinerea .
J Appl Glycosci (1999), 72, 2025
|
|
9JB2
 
 | |
9JAZ
 
 | |
9JB0
 
 | |
9JB1
 
 | |
5B6S
 
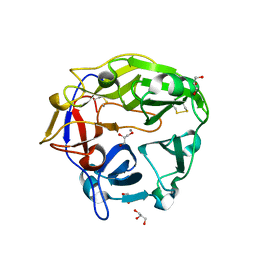 | | Catalytic domain of Coprinopsis cinerea GH62 alpha-L-arabinofuranosidase | | Descriptor: | CALCIUM ION, GLYCEROL, Glycosyl hydrolase family 62 protein | | Authors: | Tonozuka, T. | | Deposit date: | 2016-06-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Catalytic Domain of alpha-L-Arabinofuranosidase from Coprinopsis cinerea, CcAbf62A, Provides Insights into Structure-Function Relationships in Glycoside Hydrolase Family 62
Appl. Biochem. Biotechnol., 181, 2017
|
|
7XOI
 
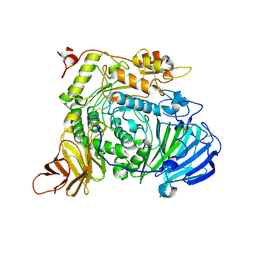 | | Aspergillus sojae alpha-glucosidase AsojAgdL in complex with trehalose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proteolytic processing of Aspergillus sojae alpha-glucosidase L with strong transglucosylation activity.
J.Struct.Biol., 214, 2022
|
|
