7DW6
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp15|16 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp15/16 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp9/10 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVX
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp6|7 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp6/7 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVW
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp5|6 peptidyl substrate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, nsp5/6 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6AJH
 
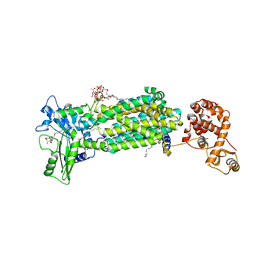 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with AU1235 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1-(2-adamantyl)-3-[2,3,4-tris(fluoranyl)phenyl]urea, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
8YZK
 
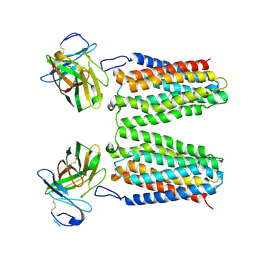 | | Orphan receptor GPRC5D in complex with scFv150-18 | | Descriptor: | Soluble cytochrome b562,G-protein coupled receptor family C group 5 member D, scFv | | Authors: | Yan, P, Lin, X, Xu, F. | | Deposit date: | 2024-04-07 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The binding mechanism of an anti-multiple myeloma antibody to the human GPRC5D homodimer.
Nat Commun, 15, 2024
|
|
8UV0
 
 | |
8GRY
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with rat ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-09-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
6BH3
 
 | |
6BGW
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(4,4-difluoropiperidin-1-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound N41) | | Descriptor: | 2-{(S)-(2-chlorophenyl)[2-(4,4-difluoropiperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGY
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(1-methylpyrrolidin-2-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound 46) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(S)-(2-chlorophenyl){2-[(2R)-1-methylpyrrolidin-2-yl]ethoxy}methyl]-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGX
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)((4,4-difluorocyclohexyl)methoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound N42) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[(4,4-difluorocyclohexyl)methoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
8H08
 
 | |
8H07
 
 | |
4J51
 
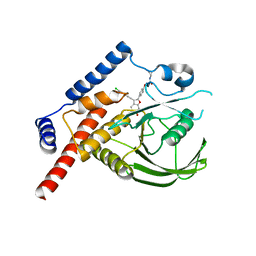 | |
8H06
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-09-28 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
8H5C
 
 | | Structure of SARS-CoV-2 Omicron BA.2.75 RBD in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Bai, B, Liu, K.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
6BH2
 
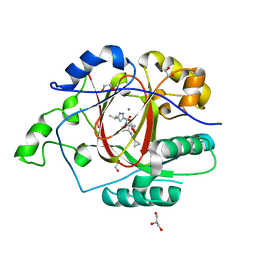 | |
6BH1
 
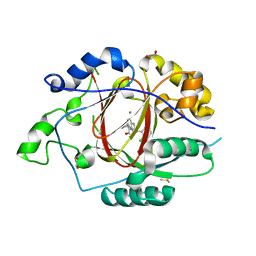 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR (S)-2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N52) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH4
 
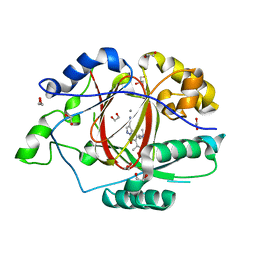 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 5-(1-(tert-butyl)-1H-pyrazol-4-yl)-6-isopropyl-7-oxo-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile (Compound N75/CPI-48) | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-tert-butyl-1H-pyrazol-4-yl)-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH0
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR (R)-2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N51) | | Descriptor: | 2-{(R)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, Lysine-specific demethylase 5A, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGZ
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(1-methyl-1H-imidazol-2-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N47) | | Descriptor: | 2-{(S)-(2-chlorophenyl)[2-(1-methyl-1H-imidazol-2-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, Lysine-specific demethylase 5A, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGU
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(propoxy)methyl)-1H-pyrrolo[3,2-b]pyridine (Compound N9) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(R)-(2-chlorophenyl)(propoxy)methyl]-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGV
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N40) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH5
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(3-(piperidin-1-yl)propoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N48) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[3-(piperidin-1-yl)propoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
